Introduction
Innate immunity is the human body’s first protection for the immune system against cellular stress and harmful incursions by generating an adaptive immune response. Various molecules are vital to supporting the immune signaling pathway, and Stimulator of Interferon Genes (STING), a homodimer transmembrane protein associated with the endoplasmic reticulum (ER), is a crucial constituent in DNA-moderated innate immunity (Ishikawa & Barber, 2008; Ishikawa et al., 2009). STING, located in the ER, is encoded by the human transmembrane protein 173 (TMEM173) and is critical for regulating the transcription of various host defense genes (Patel & Jin, 2018; Barber, 2015). STING operates in a similar way to a biological alarm system, particularly in identifying foreign DNA within the cell, which may indicate the presence of bacteria or viruses (Ishikawa et al., 2009; Gratz et al., 2011; Barker et al., 2013; Ma et al., 2015; Schoggins et al., 2015). When foreign DNA is detected, STING triggers the release of interferons, which are signaling molecules that activate immunological and antiviral responses (Bridgeman et al., 2015; Bai & Liu, 2019). STING can additionally interact with other immunological signaling molecules and cellular events, including autophagy and senescence, to enhance the reaction (Watson et al., 2015; Yang et al., 2017; Wan et al., 2020).
As STING plays an essential part in the immune response, its structure reflects its crucial role. Three domains construct STING: an N-terminal transmembrane domain (TMD, residues 1-154), a dimerization and ligand binding domain (LBD, residues 155-342), and a C-terminal tail (CTT, residues 343-379) (Tsuchiya et al., 2016; Shang et al., 2019; Zhang et al., 2019). TMD is made up of four transmembrane helices that provide proper binding of STING to the ER/Golgi membrane. At the same time, LBD binds to cyclic dinucleotides (CDNs) and functions as a sensor, and this results in a dimer composed of two STING proteins, initiating signaling cascades that regulate the immune responses (Tsuchiya et al., 2016; Shang et al., 2019; Zhang et al., 2019; Hussain et al., 2022). Located in the cytoplasm, the CTT domain is in charge of interacting with other signaling proteins, regulating STING’s stability and activity, and thereby controlling the strength and duration of the immune signaling response (Chen et al., 2016). This clearly defined structural organization enables STING to efficiently detect cytosolic DNA and proceed to trigger appropriate immune responses, hence central to innate immunity.
As a part of the cGAS-cGAMP-STING pathway, STING plays an essential role in the innate immune system, briefly explained in Figure 1. While DNA is typically confined to the nucleus and mitochondria in healthy cells, extraordinary conditions such as stress or damage can cause either mitochondrial or nuclear DNA to be released into the cytosol, triggering the cGAS-cGAMP-STING pathway as a danger signal (Sun et al., 2013). In other situations, the cGAS-STING pathway detects and responds to foreign DNA and RNA in the cytoplasm. As a STING-dependent cytoplasmic DNA receptor, cGAS is a well-established example of a pattern recognition receptor (PRR), capable of identifying and reacting to pathogen-derived and self-DNA or RNA (Yang et al., 2017).
Figure 1. cGAS-STING signaling pathway mediated by cytosolic DNA sensing. Created with Biorender.com.
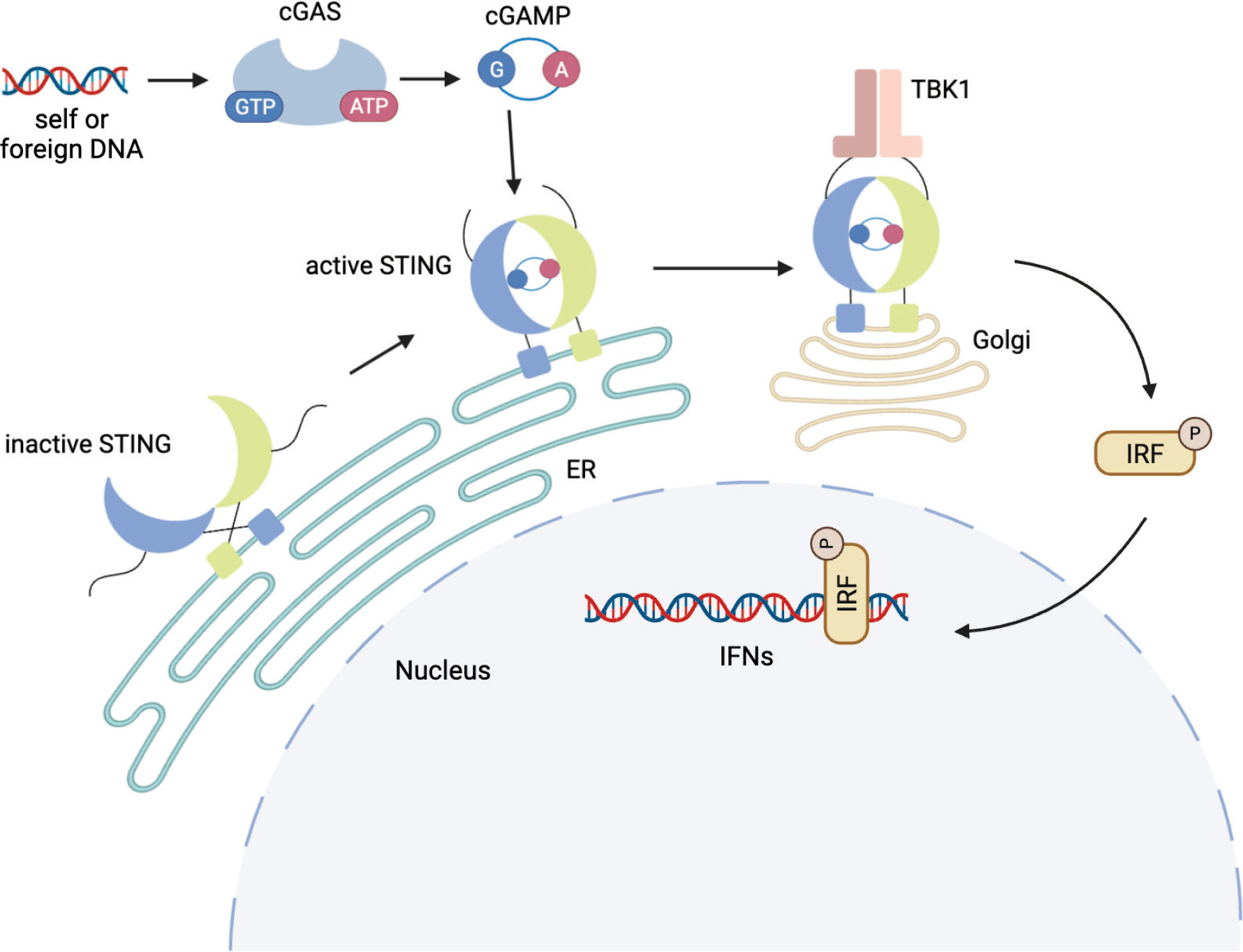
Upon DNA detection, cGAS binds to it and gets activated, which causes it to produce a signaling molecule, cGAMP. This molecule helps trigger the body’s immune response, thus activating STING. This leads to structural changes in STING, causing it to release its CTT loop, thus triggering polymerization (Smith, 2021).
Compared to the inactive structure of STING, a more structurally organized CTT loop domain is seen in the cGAMP-bound structure. Temporary β-sheet formations in the CTT loop, primarily between residues 348-350 and 362-364, are present and assist in maintaining the proper confirmation necessary for the STING protein to function (Al-Masri et al., 2021). The hydrogen bond interactions between L364-T348 and E362-A350 residues observed only in the active structure and among the residues of the temporary β-sheets (Figure 2) promote the overall stability of STING. The conserved serine residues (S355, S358, and S366) located on the CTT structure serve an essential role in STING’s ability to activate the downstream signaling (Tanaka & Chen, 2012; Tsuchiya et al., 2016). Following STING activation, these serine residues are anticipated to undergo phosphorylation by TBK1 (Tanaka & Chen, 2012; Tsuchiya et al., 2016).
Figure 2. The residues known to be part of the temporary beta sheet.
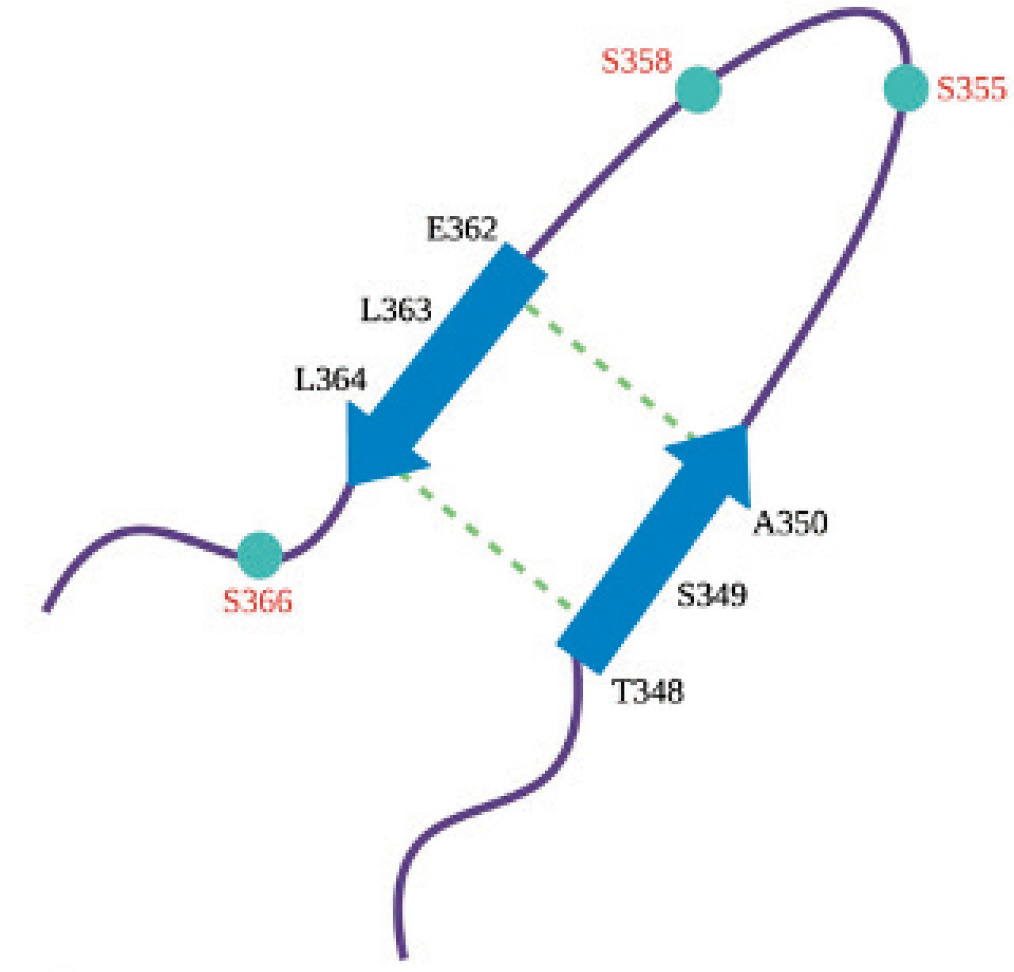
These β-sheet formations are crucial in the complex formation of TBK1 and STING, as they create the surfaces necessary for the CTT loop to interact with the proteins involved in STING signaling pathway activation and interferon production while also ensuring its correct packaging in the oligomers required for pathway activation. Given that CTT-protein interactions are fundamental for STING’s role in activating numerous immune pathways, identifying these interactions could potentially be essential for developing therapies that alter STING activity for the treatment of various diseases.
The structure and function of the STING’s CTT domain have yet to be clarified due to the inadequacy of a solved crystal structure, restricting the current knowledge of its presence in STING-TBK1 complex formation and modulation of STING-mediated signaling pathways. There are various structural studies of STING TBK1 interactions, as STING has been the focus of various studies, each contributing to a collective understanding. A comprehensive X-ray and computational study on mutations altering STING-ligand interactions has provided crucial insights for the design of STING agonists (Vavřina et al., 2021). Similarly, research on STING-TBK1 interaction and the mechanism of STING has uncovered numerous novel aspects, thanks to the collaborative efforts of many researchers. For instance, a previous study revealed that STING in the APO form is a cell oligomer (Liu et al., 2023). Another study found that Zebrafish STING possesses a unique C-terminal motif (DPVETTDY) responsible for robust NF-κB activity. When transplanted, this motif can enhance NF-κB signaling in mammalian STING, although TRAF6-binding motifs are conserved only in ray-finned fish and not in mammals (de Oliveira Mann et al., 2019). Regarding TBK1 and STING interaction, it was found that the release of CTT occurs only upon activation. However, details of how the CTT of STING captures TBK1 are missing (Zhang et al., 2022). In a study focusing on the role of the CTT loop for STING-TBK1 binding, molecular dynamics simulations of STING with the CTT loop in both ligand-bound and unbound forms were performed to understand the impact of the potent ligand cGAMP on STING and the effect of binding. The study showed that the binding of the cGAMP to STING induces local structural ordering in the CTT, which was essential for the induction of IFN-β production. The results also showed that the closed-lid conformation is only maintained in the cGAMP-bound state (Tsuchiya et al., 2016). In order to further analyze the CTT interactions, the loop modeling of the CTT domain in both the active and inactive STING conformations has been studied in the absence of cGAMP, where the modeled inactive and active STING structure with a detailed loop modeling step for CTT has been docked to TBK1 to observe the differences in binding modes (Al-Masri et al., 2021).
To understand the mechanism of the CTT loop in more detail, especially comparing the inactive and active conformations in the absence of cGAMP, this study focuses on the best models taken from the previous study and further analyzes them with molecular dynamics (MD). In the previous study by Al-Masri, MODELLER (Webb & Sali, 2016) was used to perform loop modeling of the CTT domain in inactive and active STING forms (Al-Masri et al., 2021). The modeled structures were later aligned and docked with TBK1 to analyze the change in binding interactions between the two states. After comparing the resulting STING poses with cryo-EM structures, the most compatible models were identified for further examination. Here in this study, the homology models for inactive and active STING structures are used in molecular dynamics simulations to analyze the effects of solvation and equilibration on the dynamics of the CTT loop structure.
Materials and methods
Structure preparation and system generation
MODELLER was used to form the structures (Webb & Sali, 2016) from the collected data from our previous study of the inactive and active structures of the human STING-TBK1 complex (Al-Masri et al., 2021). Active STING (PDB code: 4LOI) and inactive STING (PDB code: 4EMT) (Gao et al. 2013; Shu et al., 2012). There are two big loop structures modelled, which are missing in PDB deposited structures. These loop segments are modelled in the MODELLER software loop modeling step.
For the MD simulations performed in this study, inactive and active STING structures were extracted from the STING-TBK1 complex structures. Each STING structure contains a total of 448 amino acid residues, consisting of two chains, A and B. By using the AutoPSF Builder Plugin of VMD, protein structure files (PSF) of both inactive and active STING are generated using the topology file ‘ top_all36_prot’ from CHARMM (Brooks et al., 1983; Huang & MacKerell, 2013; Humphrey et al., 1996). Both STING structures were embedded in the TIP3P water box scaled 1 +15 Å from the molecule with the “solvate plugin” in the VMD, followed by system neutralization with Na+ and Cl- ions to create the final 0.15 M NaCl concentration (Humphrey et al., 1996). In the end, approximately 190,000 atoms were present in the systems with the box dimensions of 157.8 x 146 x 84.3 Å (Ångström).
Molecular dynamics simulations
NAMD was used for the water box-solvated inactive and active STING system. For each structure, two independent MD simulations have been carried out (Essiz et al., 2021; Phillips et al., 2020). The CHARMM parameters “par_all36_prot.prm” and “par_water_ions.prm” (Pastor et al., 1988) were used with a non-bonded van der Waals (vdW) cutoff of 12 Å. Following a 2500-step system minimization, production dynamics simulations were conducted at constant pressure (1 atm) and temperature (310 K) using a Langevin thermostat and barostat. All simulations were run for 320 ns with a time step of 2 fs, where the last 250 ns after the structures reached equilibrium were used for analysis of both first and second runs.
Trajectory analysis
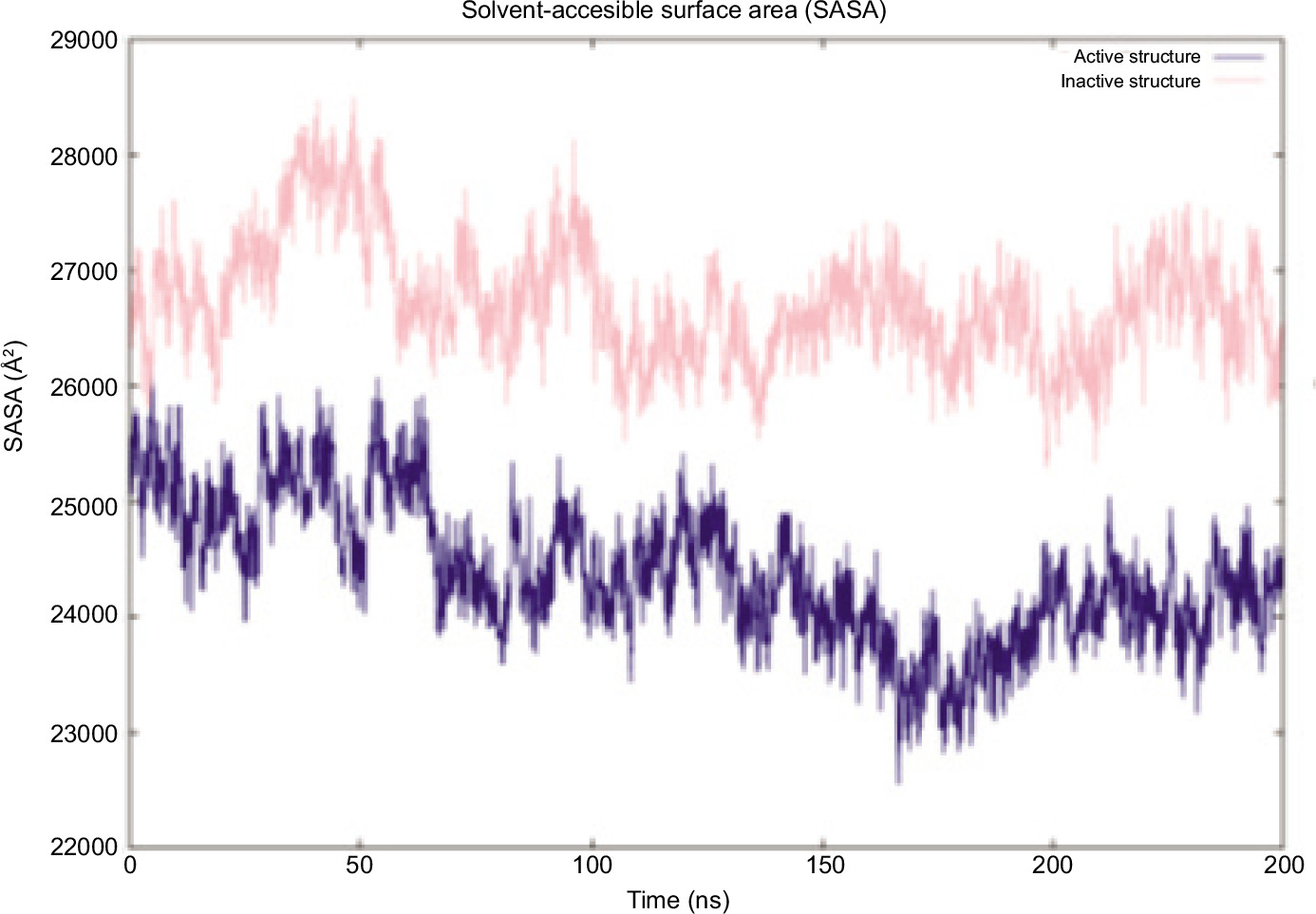
After the simulations were done for both inactive and active systems, the trajectories were analyzed to assess how the CTT loop of the STING protein behaves during the simulations for both runs. This totals to a 640 ns simulation for each structure. First, the Root Mean Square Deviation (RMSD) was calculated individually for the systems to determine if the simulation had reached equilibrium; then, the Root Mean Square Fluctuation (RMSF) was calculated to gain information about the mobility of individual residues within the molecule, the radius of gyration (Rg) was used as a metric for analyzing a molecule’s overall compactness and dynamic behavior, and finally, Solvent-Accessible Surface Area (SASA) was calculated and compared to second run (can be found in supplementary files).
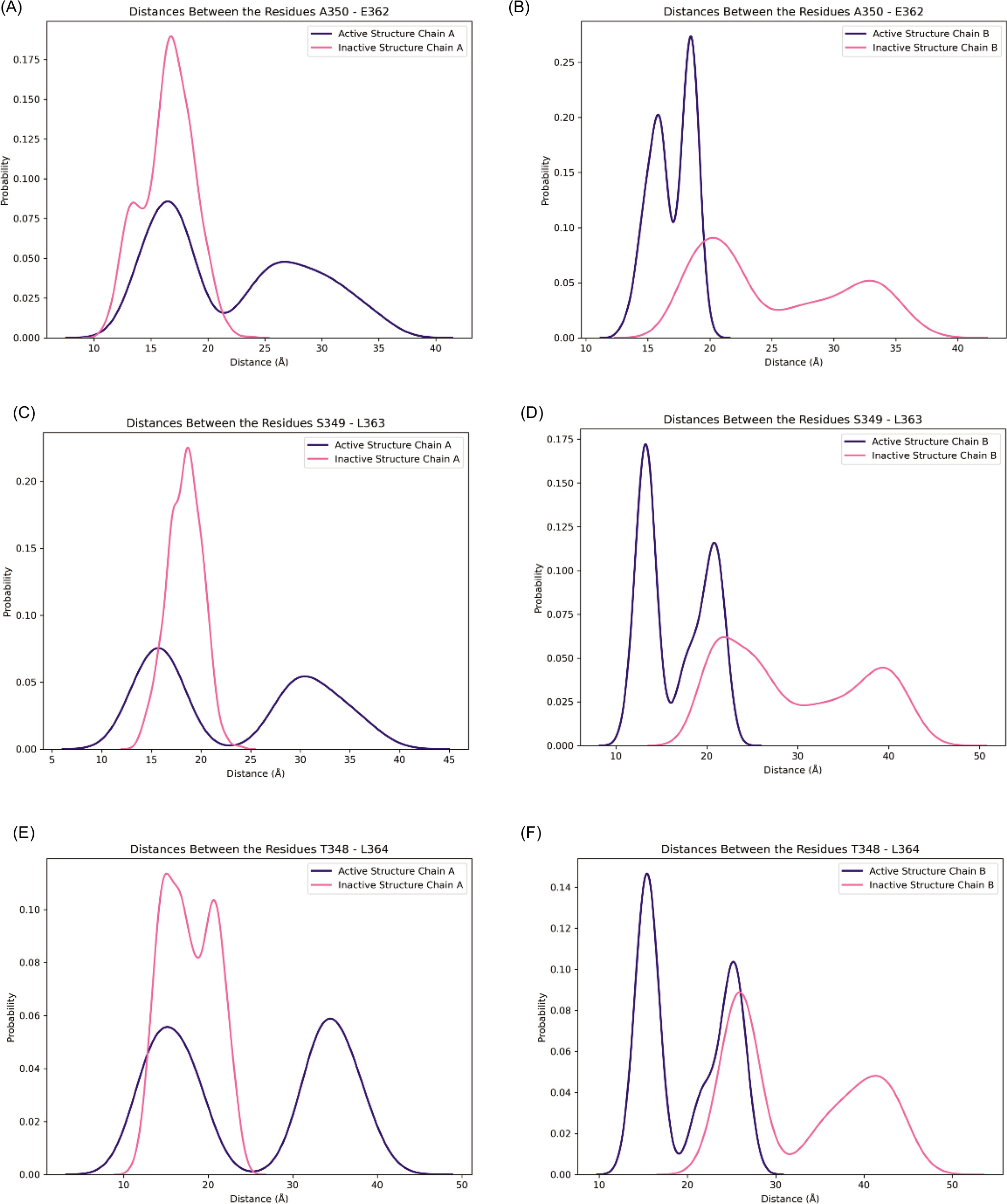
Along with these analyses, hydrogen bond analysis was performed to observe the system stability and dynamics. The distance between the two ends of the CTT loops of both inactive and active STING systems was measured to examine the movement of the loops. An additional measurement of the distance between certain amino acids (E362-A350, L363-S349, L364-T348) was conducted to observe the β-sheet formation on the CTT loop as average values of the first and second runs.
Secondary structure analysis
Secondary structure analysis was carried out for the protein at the beginning of the simulation and at 320 ns, using the DSSP (Dictionary of Secondary Structure in Proteins) tool (Touw et al., 2015) and VMD for visualization. For the hydrogen bonds, the donor-acceptor distance cutoff has been set as 3.0 Å, and the angle cutoff as 20 degrees.
Results and discussion
In a previous MD study, which also studied CTT loop structure (Tsuchiya et al., 2016), the authors observed a β-sheet formation in the c-GAMP-bound structure. In this study, the c-GAMP in the active structure is absent, but the protein stays in the stable active conformation. First, the structures at the beginning and end of the simulations are visually inspected in Figure 3. Figure 3A displays the inactive (pink) and active (indigo) states at the start of the simulation when the CTT loops are not equilibrated. At the end of 320 nanoseconds in both structures, the CTT loop adopts a more compact form (Figure 3B).
Figure 3. (A) The inactive (pink) and active (indigo) structures at the beginning of the simulations. (B) The CTT loop comes towards the lid when the simulation is at 320 ns.
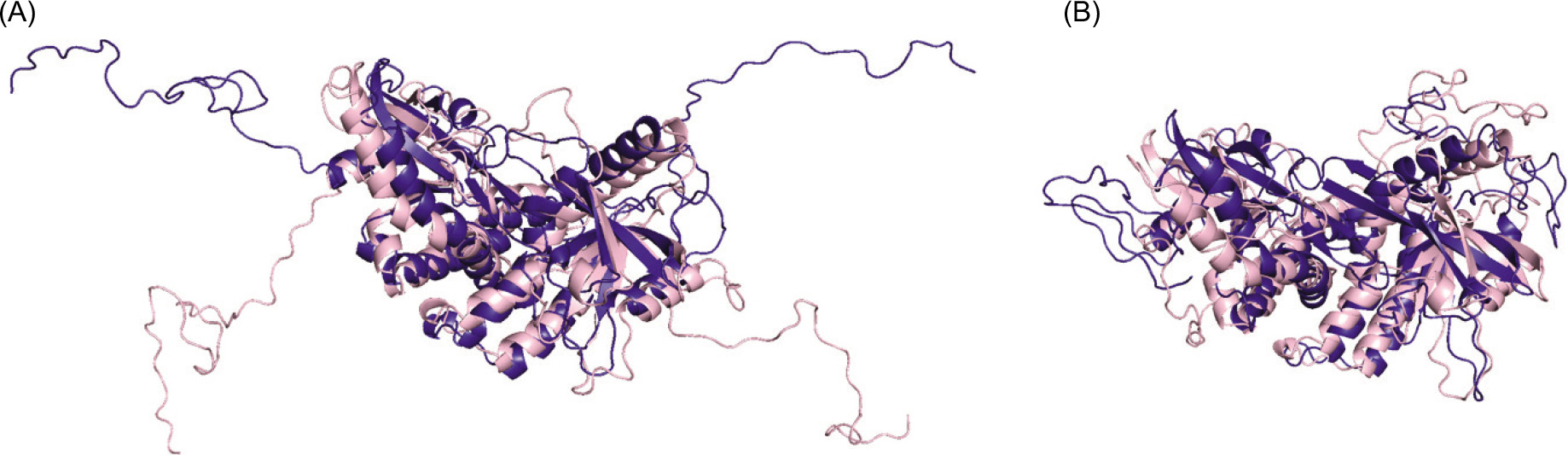
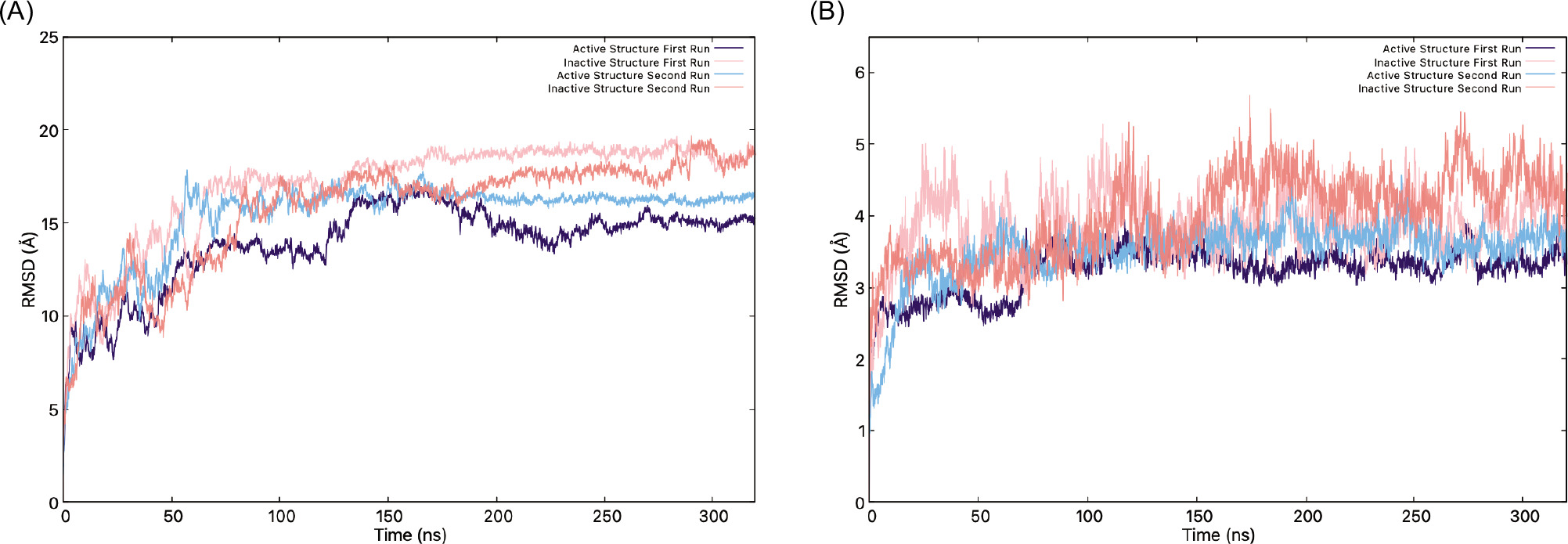
Figure 4 displays the RMSD of the protein with and without the CTT loop structure through the simulation. The initial 70 ns of the trajectory were excluded from subsequent analyses because the system had not yet reached structural equilibrium. Equilibration was assessed by monitoring the root- mean-square deviation (RMSD) of the protein backbone relative to the initial structure for all runs; the average RMSD values showed large fluctuations during this period, especially active structure in the first run, indicating that equilibrium was achieved beyond 70 ns. The RMSD graphs show that STING’s inactive and active states exhibit distinct structural behaviors, which influence their binding ability to the TBK1 protein. When the whole protein, including the CTT loops, is analyzed, the RMSD graph (Figure 4A) shows higher RMSD values in an inactive state. Meanwhile, the active state, even without the presence of c-GAMP, shows relatively lower RMSD values, suggesting a more rigid and stable structure. When the CTT is excluded (Figure 4B), both states display much lower RMSD values, highlighting the contribution of the CTT loop to the flexibility of the STING. The RMSD values range between 2 and 5 Å without the CTT loop, and the inactive state, especially in the second replica of the MD simulation, becomes much less stable than the active state with the loop. Thus, the inactive structure seems to have more flexibility compared to the active structure (please note the fluctuation amplitudes in the inactive RMSD graph for the inactive structure).
Figure 4. (A) RMSD graphs of the inactive (pink) and active (indigo) structures, (B) RMSD graphs without the CTT.
The RMSF analysis (Figure 5A and Figure S1 for the second replicas) reveals distinct differences in the flexibility of the inactive and active states. The inactive STING exhibits significantly higher fluctuations in around residues 180-230 and 400-450, both CTT loop regions. The peaks exceed 14 Å, suggesting highly dynamic and disordered segments for both chains. Meanwhile, the active structure maintains lower fluctuations during the simulation, which might indicate a more stable conformation than the inactive state. Even without the c-GAMP present, the active state preserves a well-formed and compact structure. The fluctuations increase a lot around CTT loop residues (180-230 and 400-450), especially for chain A, which might suggest that chain A’s CTT loop is the one binding to TBK1.
Figure 5. (A) RMSF graphs of the inactive (pink) and active (indigo) structures, (B) Rg values of STING through the simulation.
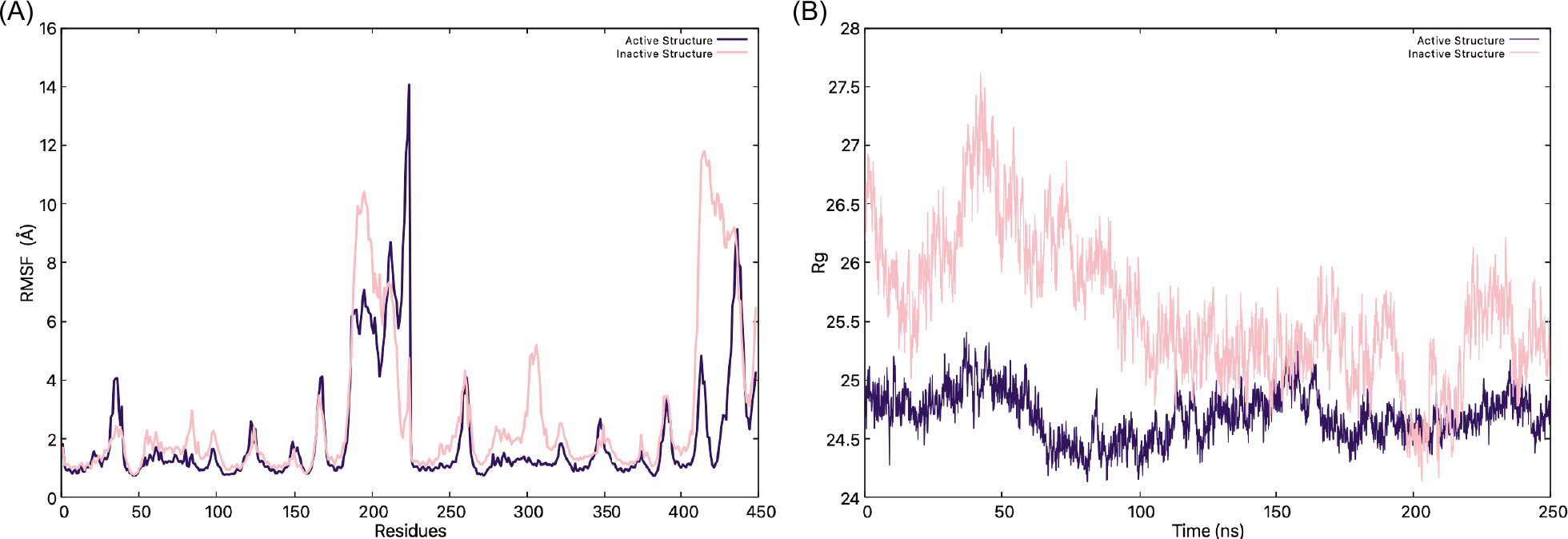
When the Rg graph is analyzed (Figure 5B and Figure S2), the findings further highlight the structural distinctions between the two states, as the inactive state consistently maintains a higher Rg value of 26 Å to 27.5 Å compared to the active state’s 24.5 Å to 25 Å, which indicates that it might adopt a more expanded and flexible conformation than the active state. The fluctuations in Rg over time suggest that the inactive STING undergoes continuous structural adjustments, while the active state is more compact with or without the c-GAMP; thus, the CTT loops must come toward the center of the protein. These results suggest that transitioning from an inactive, flexible state to a more compact and stable active state is crucial for binding STING to TBK1.
The Solvent Accessible Surface Area (SASA) over time for both the inactive and active structures is presented in Figure 6 (Figure S3 for second runs). The inactive STING exhibits a higher SASA compared to the active state throughout the simulation, indicating that it adopts a more expanded conformation with greater solvent exposure. This finding is particularly significant as it suggests that the active state, mainly because it has a lid over c-GAMP, remains impacted even without the c-GAMP. The steady decline in SASA over time suggests that both structures become more compact as the simulation progresses. However, the inactive state remains significantly more exposed, as confirmed by Rg analysis, which shows a more extended conformation in the inactive state. The binding of c-GAMP induces the necessary structural changes for the protein to bind to TBK1.
Figure 6. SASA graph of inactive (pink) and active (blue) states.
For both inactive and active states of STING, the CTT loops move closer toward the center (Figure 3). In a previous MD study, a β-sheet formation was observed in the CTT loop of the c-GAMP-bound structure (Tsuchiya et al., 2016). The existence of similar secondary structure formations was checked in the simulations. Furthermore, in the same study, hydrogen bonding between T348-L364, A350-E362, and S349-L363 pairs in the CTT loop was observed, which was again checked in this study.
In this study, a secondary structure formation was observed only for the active structure. In one replica of a 320 ns MD simulation of the active structure, chain A had a small alpha-helix, and in the second replica of a 320 ns MD simulation of the active structure, chain B displayed the same type of helix formation (Figure 7A and 7B). Meanwhile, no secondary structure was formed in the inactive structure (Figure 7C and 7D). To further analyze the pair of amino acids that are responsible for hydrogen bonding, the distance fluctuations of the hydrogen bonding amino acids in CTT have been checked during the last 50 ns of the simulations and are shown in Figure 8. These values were collected for two replicas of each structure; thus, they showed a total of 100 ns equilibrium fluctuations. The active structure samples shorter distances in chain B, while the active structure displays two peaks with shorter distances and longer distances in chain A. Although two chains of the protein display different patterns, the loop samples have shorter, more compact distances in the active structure.
Figure 7. Representation of the CTT tail and the residues (shown as licorice) that align the temporary β-sheets in STING at the end of 320 ns for the first and second runs. The protein is colored by secondary structure, where white represents coils, cyan represents turns, pink represents bridge betas, and purple represents α helices. (A) Active structure chain (A, B) Active structure chain (B, C) Inactive structure chain (A, and D) Inactive structure chain B.
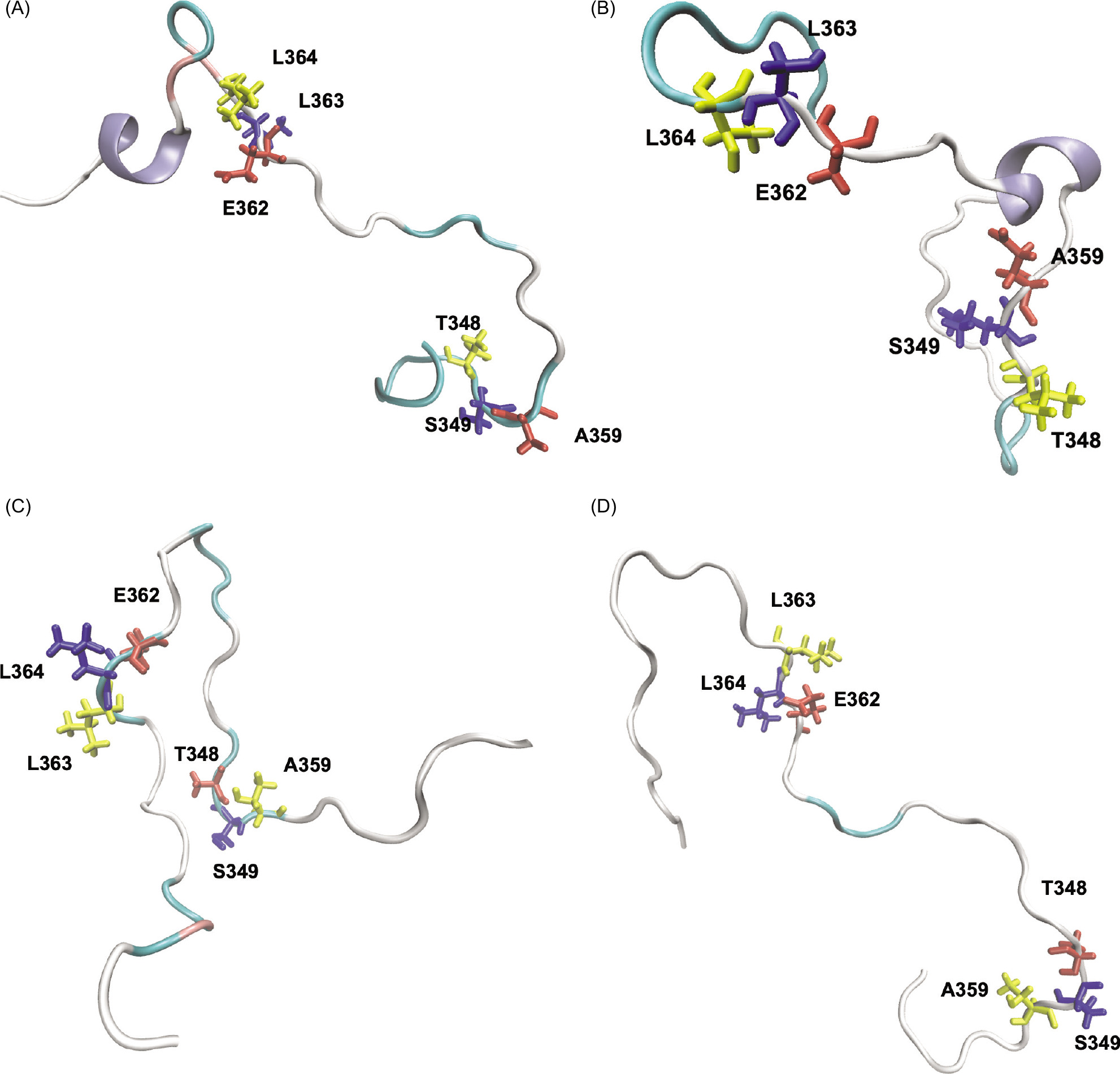
Figure 8. β-sheet forming pairs in the last 50 ns of the first and second run of simulations: Distance distribution of E350-A362, S349- L363, and T348- L364 for chain A (A, C, E) and chain B (B, D, F) are shown in pink for inactive structure and blue for active structure.
Finally, a secondary structure analysis has been carried out for the protein at the beginning and end of the simulations for the whole trajectory of 320 ns, and the average values are collected from the first and second runs. The DSSP analysis (Table 1) shows that the percentage of β-sheets varies more significantly, from 16.27% to 21.32%, with the active structure having the highest β-sheet content. In contrast, the inactive structure does not have a significant difference. Meanwhile, the coil content of the active structure is relatively lower than the inactive structure.
Table 1. DSSP analysis results.
| Structure | Helix | β-sheets | Coil |
|---|---|---|---|
| Inactive structure at 0 ns | 35.49% | 16.41% | 27.01% |
| Inactive structure at 320 ns | 36.61% | 16.27% | 27.35% |
| Active structure at 0 ns | 34.16% | 21.32% | 25.12% |
| Active structure at 320 ns | 35.71% | 20.09% | 23.11% |
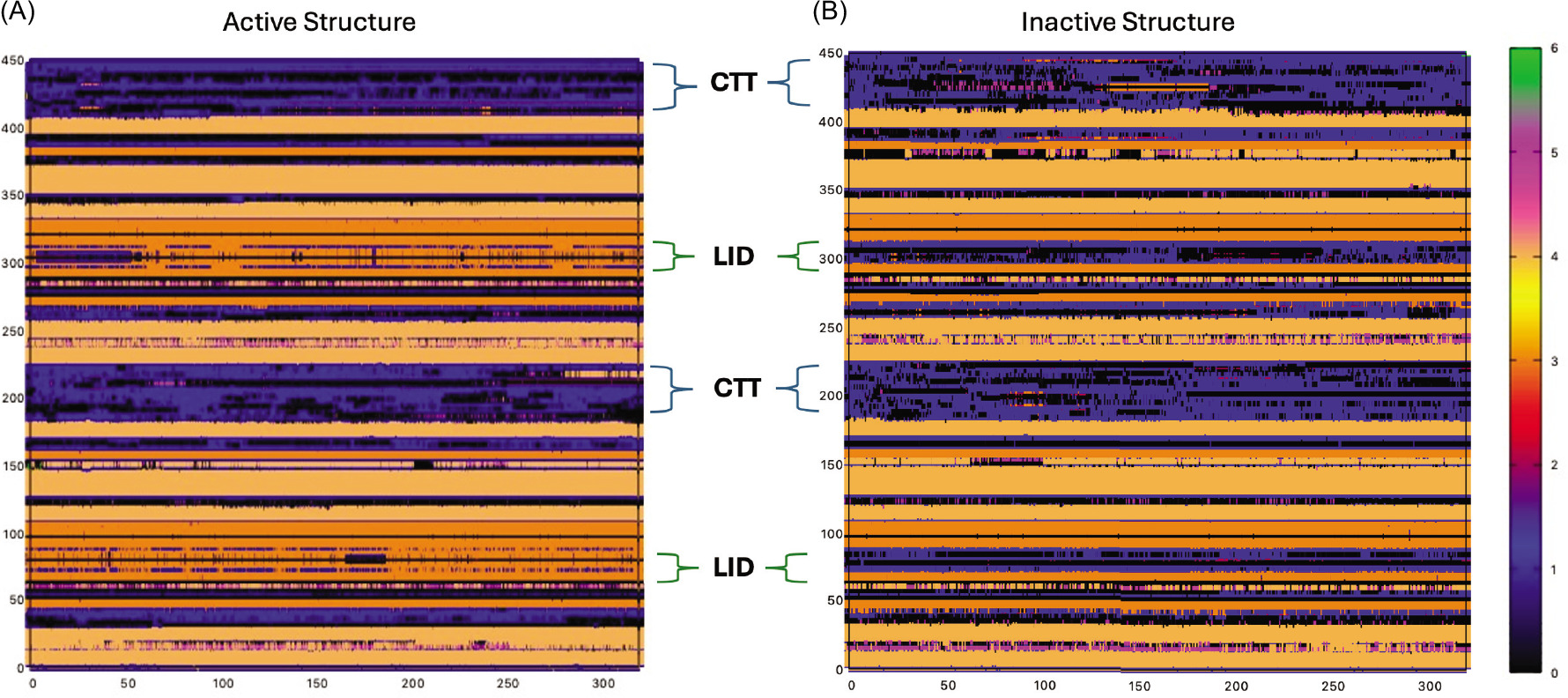
In Figure 9, the same type of secondary structure analysis has been carried out (second run analyses can be found in Figure S4). These plots display secondary structure along the different protein domains along the trajectory. The lid of the active structure accounts for the majority of the β-sheet percentage of the protein (Figure 9, please note the color change to orange in the lid domain in the active structure). CTT loops of active and inactive states also show a noticeable difference. While the CTT of chain B in the active state remains as coils, chain A starts to become α-helices. In the inactive state, there are some frames in which the coils become either helices or β-sheets; however, at the end of the simulation, both loops stay as coils.
Figure 9. Secondary Structures of active (left) and inactive (right) states. Dark Orange: β-sheet / Light Orange: α-helix/Blue: Coil/Magenta: 3-10 Helix.
Conclusions
In this study, two CTT loop structures were added to each chain of STING. And 320 ns MD simulations with two replicas for active and inactive forms of the protein were obtained. The final trajectory totaling 640 ns has been analyzed for the differences between inactive and active structures. The results reveal significant structural differences between the active and inactive STING. In the active structure, CTT loops reach equilibrium faster, as seen from the RMSD graphs with and without CTT loops. However, loop structures behave differently even in two chains of the protein, independent of the active or inactive conformation. This is an expected situation, as the loops exhibit random behavior regardless of the structure. However, one significant common observation is that in the active structures, there is some secondary structure formation observed in the CTT loop region. This is observed in both chain A and chain B of the active structure. CTT is the segment of the STING that interacts with both downstream elements of TBK1 and IRF3. Here, the active structure’s CTT loop gets shorter by forming a secondary structure.
It has been shown that STING forms a beta sheet structure to bind to the binding groove located between the kinase domain of one TBK1 subunit. However, in this configuration, the phosphorylation site Ser366 on the STING tail is positioned too far from the active site of the TBK1 kinase domain to be phosphorylated (Zhang et al., 2019). In this study, the possibility of a second secondary structure occurred in the active structure. This may contribute to having a shorter loop structure for the active structure, which can bring this phosphorylation site closer to TBK1.
Furthermore, the active structure shows a notable increase in β-sheet content, aligning with previous studies associating enhanced β-sheet formation with active conformations. In contrast, the inactive structure maintains a higher coil content, indicating greater flexibility or disorder.
One other significant finding of this study is that although there is no ligand (c-GAMP) molecule present in the active site, the cleft formed by the interface of the two chains stayed stable during our simulations. These results suggest structural rigidity, particularly in β-sheets, is conserved in STING’s activation. At the same time, increased coil regions in the inactive form contribute to its lack of structural stability, as per previous studies. The B chain of the active structure especially comes towards the center, where the lid is located, and stays there during the simulations. Since CTT loops interact with TBK1 via this CTT loop region, a more stable loop structure will make the interaction between the two proteins more specific.