Introduction
Proteases are used in various industrial fields, such as detergents, pharmaceuticals and medicine, occupying 60% of the worldwide enzyme market (Dyer & Weiss, 2022). Proteases catalyze the hydrolysis of peptide bonds, and the hydrolyzates obtained are used in the cosmetic, food and pharmaceutical industries owing to their antioxidant, antihypertensive, antidiabetic, and antimicrobial activities (Brandelli & Daroit, 2024; Elbira et al., 2024; Sun et al., 2024). Therefore, chemical, physical and enzymatic methods have been used to obtain protein hydrolyzates from different sources (Tang et al., 2023). Due to their low cost, safety and environmentally friendly nature, enzymatic methods are preferred over chemical and physical methods (Sun et al., 2024). However, the recovery problem of soluble enzymes from reaction medium, contamination of the final product with enzyme molecules as well as stability problems in organic solvents and/or at extreme pHs and temperatures make soluble enzymes economically unviable biocatalysts in many industrial processes. Enzyme immobilization is a practical and effective method to eliminate the disadvantages of using free enzymes in industrial applications. Moreover, many enzyme properties, such as activity, selectivity, specificity, and inhibition can be tuned by immobilization. Therefore, different natural and synthetic supports have been used for the immobilization of proteases (El-Shazly et al., 2024; Katić et al., 2024; Santos et al., 2024; Ungaro et al., 2024).
The use of polyvinyl alcohol (PVA) for the immobilization of enzymes and/or proteins by entrapment has been popular in recent years because of some advantages of this technique, such as ease of immobilization, low cost, protection the 3D structure of the enzyme from pH and temperature changes, and high protein loading (Toprak et al., 2021). Fernandes et al. (2009) reported that entrapped inulinase displayed high operational stability after 20 consecutive uses at 50°C. Toprak et al. (2021) entrapped nitrilase in PVA hydrogel and showed that 80% activity recovery and 100% immobilization yield were obtained. Alagöz et al. (2022) entrapped ene reductase in PVA hydrogel and demonstrated that entrapped ene reductase had 34.4-fold higher thermal stability than free ene reductase at 30°C.
In this study, a protease derived from Bacillus sp. was successfully entrapped in polyvinyl alcohol (PVA) hydrogel, and both free and entrapped forms of the enzyme were extensively characterized. The reuse stability of PVA@protease was evaluated for casein hydrolysis in a batch reactor.
Materials and methods
Materials
Protease from Bacillus sp., casein, polyvinyl alcohol, Folin Ciocalteu’s phenol reagent, glutaraldehyde (50%, w/w), and trichloroacetic acid were purchased from Sigma-Aldrich Chemie GmbH (Germany). All other chemicals were of analytical grade.
Protease immobilization
The entrapment of protease in PVA hydrogel was performed as described by Toprak et al. (2021). Before protease immobilization, 10 g of solid PVA was added to 80 mL of distilled water, and the mixture was heated to 95°C to melt the solid PVA. The mixture was then cooled to room temperature. For the entrapment of protease, 2 mL of PVA solution and one 1 mL of protease solution (1 mg protein/mL at pH 8.0) were mixed. Subsequently, 100 μL of sample was carefully dripped onto the surface of a polystyrene plate. The plates were then placed in a ventilated evaporator for 24 hours at 5°C. The entrapped protease samples (PVA@protease) were stored at 5°C until use. The amount of protein encapsulated in PVA hydrogel was determined as described by Lowry et al. (1951). The immobilization yield and recovered activity values were calculated according to Tülek et al. (2021).
Determination of protease activity
Protease activity was measured as described by Abdella et al. (2023). A 1% (w/v) casein solution was prepared in a 50 mM Tris-HCl buffer at pH 8.0. Subsequently, 0.5 mL of the casein solution was taken and incubated in a water bath at 50 °C for 1 min. The reaction was initiated by adding 50 µL of the protease solution with a concentration of 1 mg/mL. After a 5 min reaction time, 1.5 mL of TCA (10% m/v) solution was added to terminate the reaction. The mixture was then centrifuged at 10,000 x g for 15 minutes at room temperature to separate the precipitated casein from the supernatant. The absorbance of clear supernatant was measured spectrophotometrically at 280 nm.
Optimum pH and temperature
To determine the optimum pH for both free protease and PVA@protease, activity measurements were conducted using various buffer solutions across a pH range of 5.0 to 9.0. The buffers used were 100 mM acetate (pH 5.0 and 5.5), 100 mM citrate (pH 6.0), 100 mM phosphate (pH 6.5, 7.0, 7.5 and 8.0), 100 mM Tris-HCl (pH 8.5) and 100 mM borate (pH 9.0). For each pH condition, the enzyme activity was measured, with the highest recorded activity being set as 100%, and the activities at other pH levels calculated as relative values. In addition, the optimum temperature for each protease preparation was assessed by measuring their activity over a temperature range of 40°C to 70°C, maintaining pH 8.0. Similar to the pH optimization, the highest activity observed at any temperature was designated as 100%, with all other activities expressed as relative percentages.
Thermal stability
The free protease and PVA@protease were incubated in a buffer solution without substrate at 55°C to evaluate their thermal stability over time. The residual activity of each preparation was measured at specified time intervals, with the activity at zero time considered as 100%. The residual activities were then calculated relative to this initial activity. The deactivation constant (kd), half-life time (t1/2) and stabilization factor (SF) of each preparation were determined as described by Tülek et al. (2021).
Kinetic parameters
The activity of both free protease and PVA@protease was evaluated across a range of casein concentrations (2-40 mg/mL) to determine the apparent Michaelis–Menten constant (Km) and maximum reaction velocity (Vmax) for each enzyme preparation. The measurements were performed under the optimal conditions established for each protease form. To analyze the data and estimate Km and Vmax values, a Lineweaver and Burk plot was used.
Reuse stability
Reuse stability of PVA@protease was carried out in a batch reactor (1.1x5 cm) at the determined optimum pH and temperature. After each cycle, the PVA@protease was washed with distilled water and these processes were repeated 5 times.
Results and discussion
In this study, a protease from Bacillus sp. was encapsulated in PVA hydrogels and the immobilization yield and recovered activity values were determined to be 100% and 90%, respectively. Sinha and Khare (2015) reported the immobilization yield was 15% for Bacillus sp. EMB9 protease was immobilized on silica nanoparticles by simple adsorption. When silica nanoparticles were activated by glutaraldehyde, the immobilization yield increased to 40%. Thakrar and Singh (2019) encapsulated protease from Nocardiopsis alba TATA-5 in calcium alginate and determined the immobilization yield and recovered activity values were 96.6% and 44.8%, respectively.
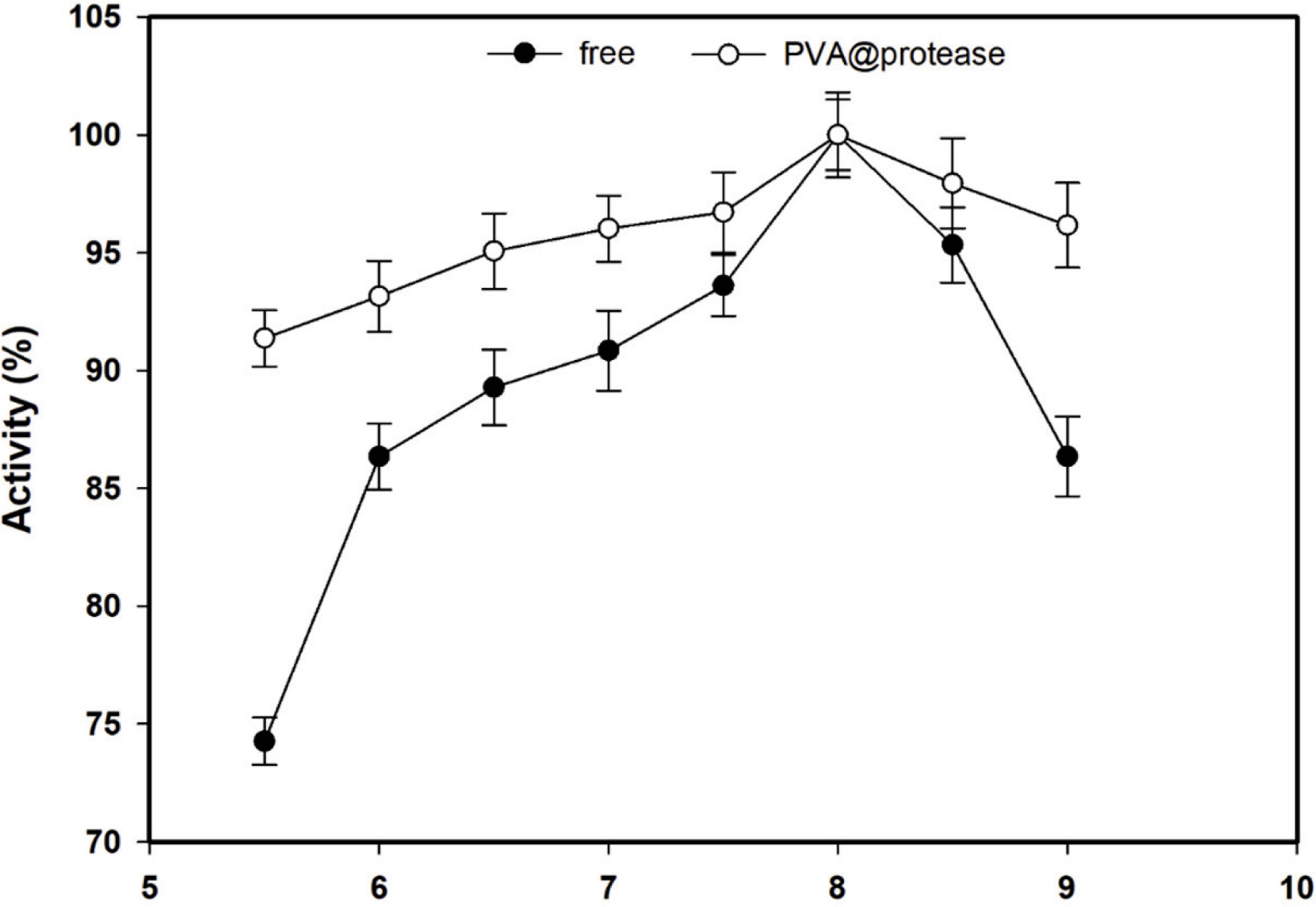
The changes in the activities of free protease and the PVA@protease with increasing pH were investigated in different buffer solutions in the pH range of 5.5-9.0 and the results are presented in Figure 1. Both proteases show their maximum activities at pH 8.0. As seen in Figure 1, the PVA@ protease exhibits higher relative activity at pH values below and above the maximum pH. These findings indicate that the stability of the protease against pH changes increases after immobilization.
Figure 1. Effect of pH on the activity of free protease and PVA@pro-tease at 55 °C.
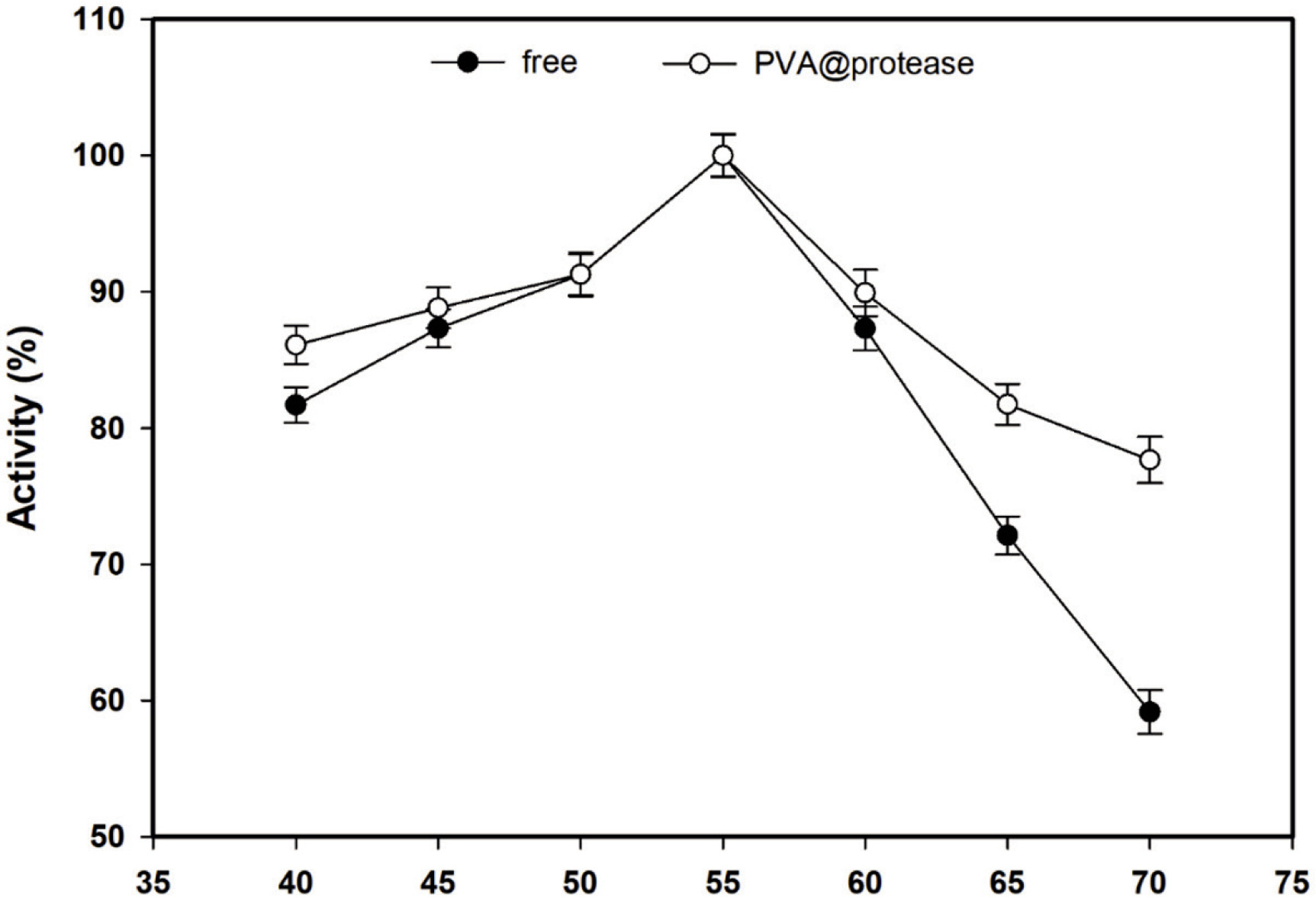
The activity change of free and immobilized protease against temperature was measured at pH 8.0 in the range of 40-70°C and the results are given in Figure 2. Both free protease and the PVA@ protease showed their maximum activity at 55°C. Free protease and the PVA@protease displayed almost similar relative activity values in the range of 40-55°C, while the PVA@protease demonstrated higher relative activity values than free protease at 65°C and 70°C. This indicates that the resistance of the PVA@protease increased to denaturation at 65°C and 70°C.
Figure 2. Effect of temperature on the activity of free protease and PVA@protease at pH 8.0.
Ferreira et al. (2003) immobilized Bacillus licheniformis protease onto 3-APTES-activated silica supports via glutaraldehyde spacer and found the optimum pH and temperature values as 8.0 and 60°C for both free and immobilized enzymes, respectively. Nakashima et al. (2006) reported the optimum pH value as 8.0 for free B. licheniformis protease. Anwar et al. (2009) immobilized B. subtilis protease in calcium alginate gel and found the optimum pH and temperature values 7.5 and 50°C for both the free and immobilized protease, respectively. Sinha and Khare (2015) covalently immobilized Bacillus sp. protease onto silica nanoparticles and determined the optimum pH and temperature as 9.0 and 55°C for both enzymes, respectively. Ramalho and de Castro (2023) determined the optimum pH and temperature values for both B. licheniformis protease immobilized on free and glutaraldehyde-modified chitosan as 9.0 and 60°C, respectively.
To determine the thermal stability of the free protease and thr PVA@protease, the remaining activities of the enzymes were measured for different incubation times at 55°C and the results are given in Figure 3. The activity of the free protease decreased linearly with increasing incubation time and its remaining activity was calculated as 19% at the end of the 24-hour incubation. The remaining activity was calculated as 72% for the PVA@protease under the same conditions. The results show that entrapping the protease in polyvinyl alcohol creates a protective effect against denaturation. The kd and t1/2 values of free protease were calculated to be 65x10-2 h-1 and 10.6 h, respectively (Table 1). These values correspond to 13x10-2 h-1 and 53.3 h for the PVA@protease. SF value was 5.0 at 55°C, showing that thermal stability of the free enzyme enhances 5.0-fold upon immobilization. Wahba (2022) indicated that B. licheniformis protease immobilized on functionalized gum tragacanth-agar (iBPR) protected 59.8% of its initial activity after 1 h incubation at 56°C, while the free protease preserved only 13.80% of its initial activity under the same conditions. Ferreira et al. (2003) reported that B. licheniformis protease immobilized on free and 3-APTES-activated silica supports via glutaraldehyde spacer lost 50% and 10% of their initial activities after 2 h incubation at 50°C, respectively. Sinha and Khare (2015) reported that free Bacillus sp. EMB9 protease lost 70% of its initial activity after 12 h incubation at 70°C, and the t1/2 value at this temperature was 3 h. However, the covalently immobilized Bacillus sp. EMB9 protease on silica nanoparticles lost 20% of its initial activity under the same conditions, and the t1/2 value was 8.8 h.
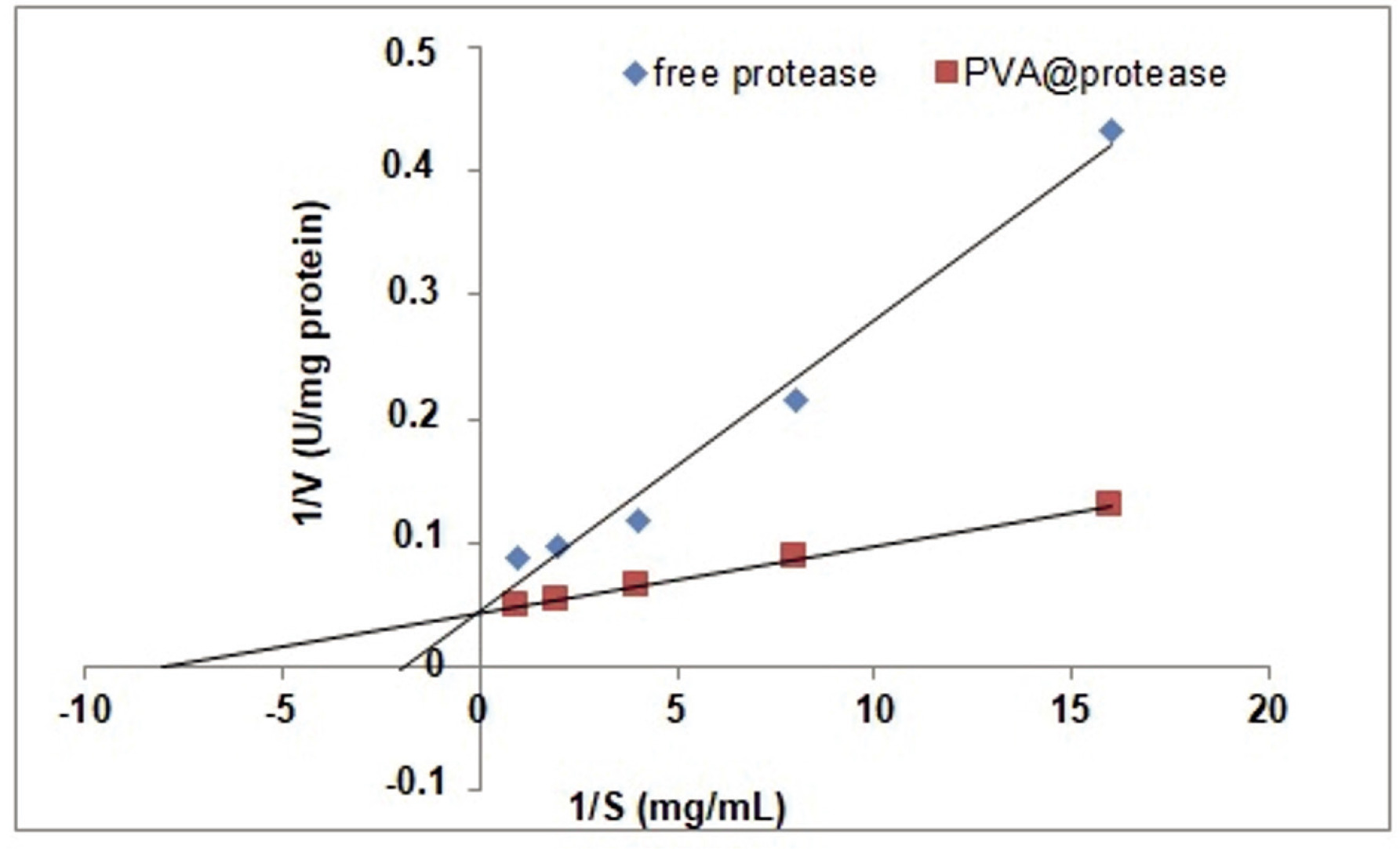
Km values for free protease and PVA@protease were estimated as 0.5 and 0.2 mg/mL for casein, respectively. This result indicates that the affinity of the enzyme to substrate increases 2.5-fold upon immobilization. Encapsulating the protease in PVA hydrogel may have indeed promoted proper orientation and therefore, the affinity of the protease to its substrate may have increased. The corresponding Vmax was determined as 23.3 and 23.8 U/mg (Figure 4).
Table 1. Thermal stability parameters of free protease and PVA@protease at pH 8.0 and 55°C.
| Enzyme form | kd (h-1) | t1/2 (h) | SF |
|---|---|---|---|
| Free protease | 65 × 10-2 | 10.6 | - |
| PVA@protease | 13x10-2 | 53.3 | 5 |
Figure 3. Thermal stability of free protease and PVA@protease at pH
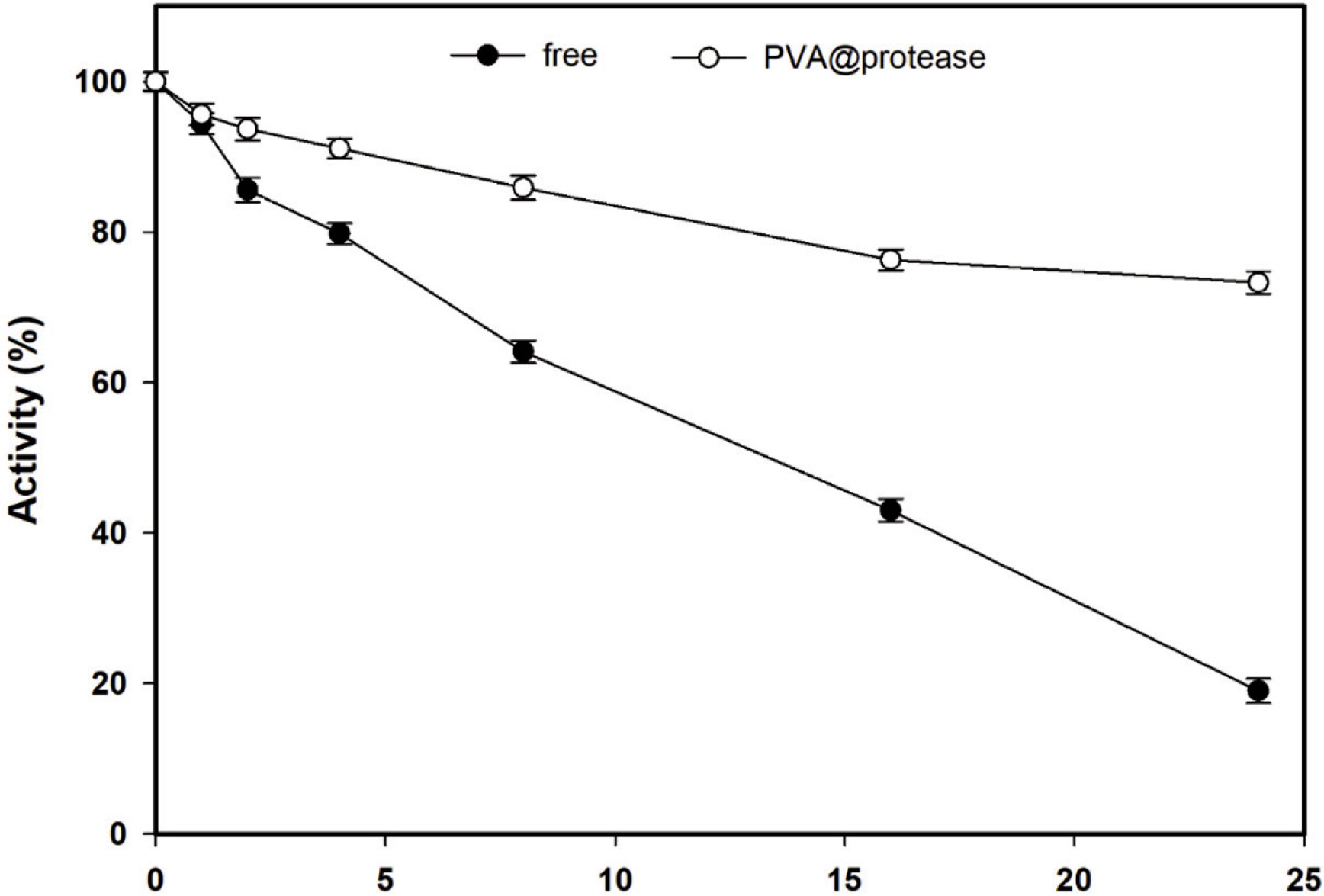
Figure 4. Lineweaver-Burk plot of free protease and PVA@protease at pH 8.0 and 55°C for different casein concentrations.
Wahba (2022) determined Km and Vmax values as 0.8 mg/mL and 7.8 U mg protein-1, respectively for free B. licheniformis protease and 1.2 mg/mL and 3 U mg protein-1, respectively for iBPR. Abdella et al. (2023) immobilized protease from B. thuringiensis on onto activated Alginate/dextrose (Alg/dex/protease) beads and reported that the Km values of the free and Alg/dex/protease was 2.56 and 5.26 mg/mL, respectively towards casein. The corresponding Vmax values were determined to be 121.95 and 149.52 U/mL/min. The Km and Vmax values of free protease from B. pumilus Y7 were 6.05 μM and 20.16 U min-1, respectively towards casein. These values correspond to 27 μM and 34.13 U min-1 for its entrapped counterpart in poly(vinylimidazole)/clay hydrogel. Adetunji and Olaniran (2023) reported the Km value of the free protease from B. aryabhattai was 2.023 mg/mL for casein, while the encapsulated protease in alginate showed a lower Km value of 1.225 mg/mL. The corresponding Vmax values were found to be 232.56 and 250 U/mL.
To evaluate the reuse stability of the immobilized protease, the enzyme was utilized in a batch reactor for casein hydrolysis across five successive cycles and the results are depicted in Figure 5. The PVA@protease retained 58% of its initial activity after 5 uses. Anwar et al. (2009) observed that a protease from Bacillus subtilis immobilized in calcium alginate gel retained only 35% of its activity after three uses. Sinha and Khare (2015) reported that a protease from Bacillus sp. EMB9, retained 75% of its initial activity after six uses when covalently immobilized on silica nanoparticles. Ramalho and de Castro (2023) reported that B. licheniformis protease immobilized on glutaraldehyde-modified chitosan retained 47% of its initial activity after 3 uses. The iBPR remained 37.6 % of its initial activity after 10 reuses (Wahba, 2022). Thakrar and Singh (2019) demonstrated that the protease encapsulated in alginate beads preserved 85% of its initial activity after 10 successive uses. Duman and Tekin (2020) immobilized protease from B. pumilus Y7 in poly(vinylimidazole)/clay hydrogel and showed the immobilized protease lost 35% of its initial activity after six cycles.
Figure 5. Reuse stability of PVA@protease.

Conclusions
In this study, a protease from Bacillus sp. was successfully immobilized in PVA hydrogels. The free and PVA@protease were then characterized. Both the protease preparations showed their optimal activities at pH 8.0 and 55°C. The residual activities of free and PVA@protease were found to be 19% and 72%, respectively, after 24 h of incubation at 55°C. The SF value was calculated to be 5, indicating that stability of the protease increased 5-fold by encapsulation in PVA hydrogel at 55°C. PVA@protease protected 58% of its initial activity after 5 uses. In conclusion, entrapment of the protease in PVA hydrogels is indeed a straightforward and effective method due to mild immobilization conditions. Because PVA is biocompatible, the PVA@protease can be used in the preparation of protein hydrolyzates for the food industry.