Introduction
Fungal infections affect more than one billion people worldwide each year, with more than 1.6 million infected cases resulting in death (Bongomin et al., 2017; Gupta et al., 2012). Among the microorganisms that cause disease in humans in the clinic, two important members of the yeast class can be ranked as Candida albicans and Candida glabrata according to their frequency of disease in humans (Calderone & Clancy, 2011). Invasive nosocomial infections are caused by Candida species, particularly in patients with impaired immune systems. Of these, Candida albicans is the most prevalent species that causes non-invasive candidiasis of the skin and mucous membranes (Tamo, 2020). But non-albicans Candida species—like C. glabrata, C. parapsilosis, C. tropicalis, C. krusei, C. lusitaniae, C. dubliniensis, and C. guilliermondii—have shown a more than 50% increase in prevalence in recent years (Seyedmousavı et al., 2015; Tamo, 2020). According to recent research, C. glabrata is more frequently isolated in cases of invasive candidiasis and is linked to a higher patient death rate. Depending on the region under investigation, C. glabrata is either the second or third most commonly isolated species of Candida (Guinea & Infection, 2014). Additionally, C. glabrata is frequently encountered in the environment, particularly on surfaces, water, soil, flowers, and leaves. After Candida albicans, it is the second most common isolated cause of candidiasis. About 15–25% of invasive clinical cases are accounted for by it (McCarty & Pappas, 2016; Pfaller & Diekema, 2007). About 40–60% of cases of invasive candidiasis caused by Candida glabrata result in substantial morbidity and mortality, possibly as a result of the bacteria's intrinsic low sensitivity to the most widely used azoles (Timmermans et al., 2018). C. glabrata is basically a yeast, but what distinguishes it from non-pathogenic yeasts used as bread and brewer's yeast are differences in the structure of the cell wall. Studies using various biochemical tests have shown that adhesin-like glycophosphatidylinositol proteins are present in higher amounts in C. glabrata than in other yeasts (Weig et al., 2004). Mass-spectrometry was then used to identify the proteins more accurately, resulting in the identification of four new adhesin-like proteins. In addition to the results, these proteins were subsequently named Awp 1,2,3,4. In addition, Epa6 from the lectin-like EPA family, which clusters with these four new adhesin-like proteins, is responsible for the gel-like biofilm layer formed by C. glabrata to adhere to human epithelial cells. (de Groot et al., 2008) In conclusion, Awp2,3,4 and Epa6, which are novel adhesin-like proteins of C. glabrata, were found only in C. glabrata strains obtained in stool test results, while Awp1 was found in strains isolated from blood. Treatment of C. glabrata infections is complicated by their inherent resistance to antifungals, especially azoles. (Mota et al., 2015) Long-term exposure to antifungal drugs for treatment and prevention of infections may be the main reason for the development of newly drug-resistant strains, according to analytical studies (Jensen et al., 2016). The virulence of C. glabrata was investigated by combining biochemical tests and mycoscopy methods, and bioinformatics studies were utilized in further studies (Enkler et al., 2016).
In this study, the crystallographic structure of AWP1 which belongs to fungus C. glabrata organism was determined as a macromolecule and the designing of new ligands as candidate drug active molecules that can show antifungal effect was aimed. While designing new drug active molecules, the study was started using the hesperetin molecule. Afterwards, derivatization studies of the molecule were carried out with functional groups added to the hesperetin molecule.
The aglycone form of the hesperedin molecule, a flavanone glycoside abundant in citrus fruits, is called hesperetin. The scientific and common names of the plants containing the hesperetin molecule and the plant parts containing the molecule are given in Table 1.
Table 1. List of Plants Containing Hesperetin Molecule.
| Plant | Common Name | Part |
|---|---|---|
| Artemisia dracunculus | Estragon | Shoot |
| Citrus paradisi | Grapefruit | Fruit |
| Citrus spp. | Citrus | Plant |
| Cynara cardunculus | Cardoon | Leaf |
| Eriodictyon californicum | Yerba santa | Resin, exudate, sap |
| Mentha aquatica | Water-mint | Plant |
| Mentha x piperita | Mentha spicata | Leaf |
Resource: U.S. Department of Agriculture, Agricultural Research Service. (1992-2016). “Dr. Duke's Phytochemical and Ethnobotanical Databases. Home Page”. (Duke, 2020)
The two-dimensional structure of the hesperetin molecule and the positions on this structure where functional groups are planned to be attached are named as R1, R2, R3, R4, R5, R6, R7, R8.
In Table 2, functional groups and ligand numbers added to the binding positions described in Figure 1 are given. Based on the skeletal structure of the Hesperetin molecule, 100 molecular modifications were performed.
Figure 1. 2D structure and derivatization positions of the Hesperetin molecule.
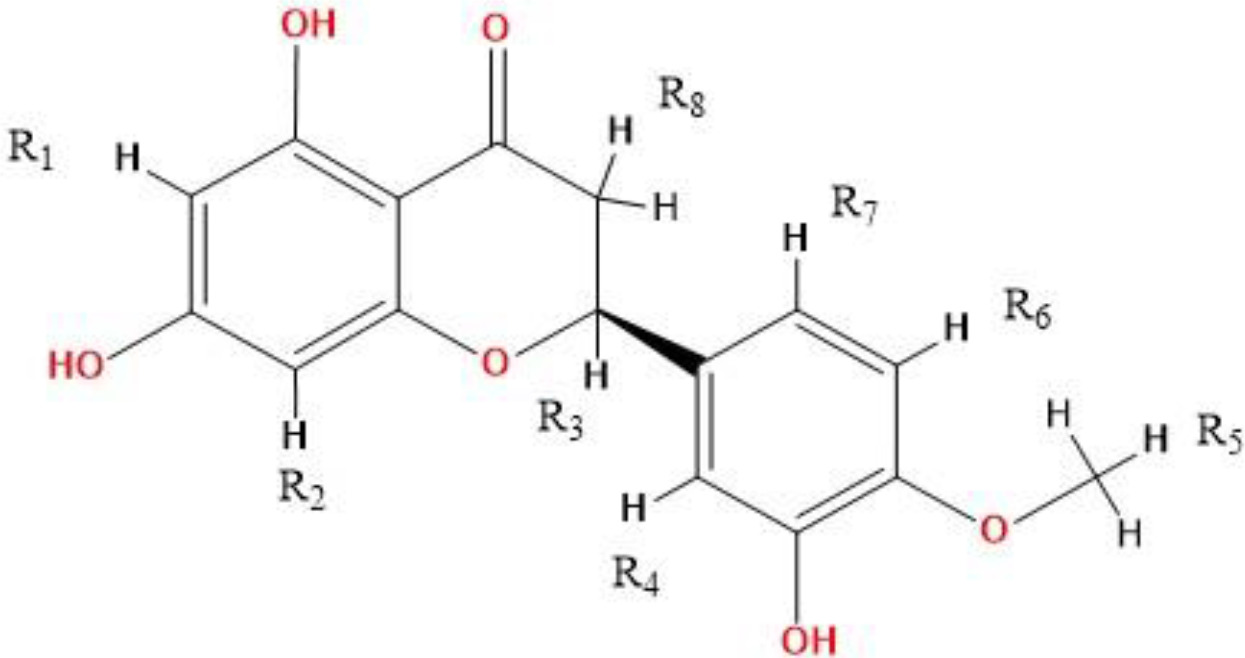
Table 2. Functional groups attached to the Hesperetin molecule and their positions.
| Ligands | R1 | R2 | R3 | R4 | R5 | R6 | R7 | R8 |
|---|---|---|---|---|---|---|---|---|
| L-1 | NH2 | |||||||
| L-2 | OH | |||||||
| L-3 | CH3 | |||||||
| L-4 | CH2F | |||||||
| L-5 | NO2 | |||||||
| L-6 | CH2OH | |||||||
| L-7 | CH3 | |||||||
| L-8 | CH2Cl | |||||||
| L-9 | CH3CH2 | |||||||
| L-10 | COOH | |||||||
| L-11 | C(CH3)3 | |||||||
| L-12 | OH | |||||||
| L-13 | OH | |||||||
| L-14 | OH | |||||||
| L-15 | OH | |||||||
| L-16 | OH | |||||||
| L-17 | OH | |||||||
| L-18 | OH | |||||||
| L-19 | CH3 | |||||||
| L-20 | CCOH | |||||||
| L-21 | CH2CH2CH2 | |||||||
| L-22 | CH2CH3 | |||||||
| L-23 | CHOHCH3 | |||||||
| L-24 | CH2OCH3 | |||||||
| L-25 | CH2C(CH3)2OH | |||||||
| L-26 | CONH2 | |||||||
| L-27 | CONH2 | |||||||
| L-28 | COCH3 | |||||||
| L-29 | C6H13 | |||||||
| L-30 | NO2 | |||||||
| L-31 | C(CH3)3 | |||||||
| L-32 | C6H5 | |||||||
| L-33 | C6H5O | |||||||
| L-34 | C6H5O2 | |||||||
| L-35 | O | |||||||
| L-36 | CH2CH2OH | |||||||
| L-37 | CH2CH2CH2OH | |||||||
| L-38 | CH2CH2CH2OH | |||||||
| L-39 | C(CH3)2OH | |||||||
| L-40 | C6H5 | |||||||
| L-41 | C6H5 | |||||||
| L-42 | C6H5 | |||||||
| L-43 | C6H5 | |||||||
| L-44 | C6H5 | |||||||
| L-45 | C6H5 | |||||||
| L-46 | CH3 | |||||||
| L-47 | CH3 | |||||||
| L-48 | CH3 | |||||||
| L-49 | CH3 | |||||||
| L-50 | NH2 | |||||||
| L-51 | NH2 | |||||||
| L-52 | NH2 | |||||||
| L-53 | NH2 | |||||||
| L-54 | NH2 | |||||||
| L-55 | NH2 | |||||||
| L-56 | C2H3O | |||||||
| L-57 | OH | CH3 | ||||||
| L-58 | OH | OH | ||||||
| L-59 | OH | O | ||||||
| L-60 | NH2 | NO2 | ||||||
| L-61 | C6H5 | O | ||||||
| L-62 | C6H5 | CH3 | ||||||
| L-63 | C6H5 | CONH2 | ||||||
| L-64 | C6H5 | CONH2 | ||||||
| L-65 | CONH2 | C(CH3)2OH | ||||||
| L-66 | CONH2 | C6H5O | ||||||
| L-67 | NH2 | C7H7OS | ||||||
| L-68 | CH2CH3 | C6H5OS | ||||||
| L-69 | OH | O | ||||||
| L-70 | NH2 | C6H3F2 | ||||||
| L-71 | C6H3Cl2 | C3H3N2 | ||||||
| L-72 | C3H3N3 | O | ||||||
| L-73 | NH2 | C6H3Cl2 | ||||||
| L-74 | NH2 | C6H3F2 | ||||||
| L-75 | C2H3FNO | O | ||||||
| L-76 | CH3 | C4H3O2S | O | |||||
| L-77 | NH2 | C(CH3)3 | O | |||||
| L-78 | C6H5 | OH | S | |||||
| L-79 | OH | C6H3F2 | ||||||
| L-80 | NH2 | C(CH3)3 | OH | |||||
| L-81 | OH | C3H5O2 | ||||||
| L-82 | NH2 | OH | C3H5O3 | |||||
| L-83 | CONH2 | C3H5O2 | ||||||
| L-84 | CH3 | C6H5O | O | |||||
| L-85 | CH3 | OH | CH2Cl | |||||
| L-86 | OH | CH2Cl | NH2 | |||||
| L-87 | CH3 | OH | CONH2 | |||||
| L-88 | C6H3Cl2 | OH | CONH2 | |||||
| L-89 | C4H7O2 | CONH2 | ||||||
| L-90 | CONH2 | OH | CF3 | |||||
| L-91 | CONH2 | OH | CF3 | |||||
| L-92 | CONH2 | OH | CCl3 | |||||
| L-93 | NH2 | CONH2 | OH | CONH2 | ||||
| L-94 | CH2OH | OH | CH3 | |||||
| L-95 | Cl | CONH2 | OH | |||||
| L-96 | CH2OH | OH | Cl | |||||
| L-97 | CONH2 | C6H5O | CCl3 | |||||
| L-98 | CONH2 | SH | OH | |||||
| L-99 | CONH2 | OH | CH2CH2OH | |||||
| L-100 | CONH2 | C6H5O | CH2CH3 |
Materials and methods
In the study, conformer distribution analysis with molecular mechanics/MMFF method and geometry optimization of the designed molecules were performed using Semi Empirical/PM6 method in Spartan'14 (Spartan ’14 V1.1.4) software. Visualization of the selected protein structure before docking studies was performed using the BIOVIA Discovery Studio software (Biovia, 2021). During these imaging processes, the water molecules in the crystal structure of the protein were deleted and the active site coordinates were selected as given in Table 3. The determined active site coordinates were also selected as the gridbox center coordinates in the Autodock Tools software (Morris et al., 2009) used in the preparation of the docking files of proteins and ligands in the continuation of the studies, and other data used for the gridbox are given in Table 3.
Table 3. Grid Box Coordinates, Size and Spacing of the protein.
| Coordinates | Grid box size | Grid space |
|---|---|---|
| x = 8.968 | x=40 | 0.374 Å |
| y = 28.866 | y=40 | |
| z = 41.670 | z=40 |
Docking studies of protein and derivatized ligand molecules were performed by running the Autodock Vina software on the command prompt (Eberhardt et al., 2021). After the docking scores were calculated, the results obtained were analyzed using the BIOVIA Discovery Studio software. In the analyses, protein-ligand interaction maps were generated and the interaction types and distances of the ligand molecule with the amino acids around the active site in the target protein structure were noted in these maps.
Results and discussion
For each modification, area, volume, molecular weight(MW), energy, EHOMO, ELUMO, polarizability (α), dipole moment(μ), logP values of these molecules were calculated using Spartan'14 software and applying the Semi-Empirical/PM6 method. The results obtained are given in Table 4. In addition to the physicochemical parameters, the Binding Energies (BE) of the molecules with the protein structure and the inhibition constants (Ki) of these molecules also calculated.
Table 4. Physicochemical parameters of hesperetin and its derivatives.
| Ligands | Area (Å2) | Volume (Å3) | MW (amu) | Energy (kcal.mol-1) | EHOMO (eV) | ELUMO (eV) | α | μ (debye) | logP | BE (kcal.mol-1) | Ki |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Fluconazole | 302.51 | 281.30 | 306.276 | -41.76 | -10.35 | -0.97 | 61.98 | 2.97 | -1.04 | -8.6 | 4.96 × 10-7 |
| Hesperetin | 301.62 | 285.56 | 302.282 | -853.19 | -8.94 | -0.62 | 62.57 | 4.70 | -3.12 | -8.7 | 4.19 × 10-7 |
| L-1 | 315.11 | 296.34 | 317.297 | -907.05 | -8.97 | -0.65 | 63.45 | 4.11 | -4.84 | -8.9 | 2.99 × 10-7 |
| L-2 | 309.3 | 292.86 | 318.281 | -1038.63 | -8.84 | -0.86 | 63.25 | 4.72 | -4.21 | -9.4 | 1.29 x710-7 |
| L-3 | 319.76 | 303.53 | 316.309 | -991.75 | -8.93 | -0.62 | 64.04 | 5.13 | -2.95 | -9.2 | 1.8 × 10-7 |
| L-4 | 324.02 | 308.54 | 334.299 | -1091.90 | -8.98 | -0.67 | 64.44 | 4.69 | -2.91 | -8.7 | 4.19 × 10-7 |
| L-5 | 327.78 | 307.07 | 347.279 | -828.18 | -9.36 | -0.74 | 64.25 | 3.10 | -4.46 | -9.3 | 1.52 × 10-7 |
| L-6 | 322.99 | 310.15 | 332.308 | -1097.20 | -9.09 | -0.57 | 64.52 | 6.35 | -4.01 | -8.7 | 4.19 × 10-7 |
| L-7 | 319.10 | 303.24 | 316.309 | -930.19 | -8.76 | -0.56 | 64.04 | 5.39 | -2.95 | -9.3 | 1.52 × 10-7 |
| L-8 | 336.28 | 318.10 | 350.754 | -944.40 | -9.16 | -0.67 | 65.18 | 4.85 | -2.41 | -9.0 | 2.53 × 10-7 |
| L-9 | 339.86 | 321.99 | 330.336 | -934.96 | -8.93 | -0.60 | 65.53 | 5.06 | -2.53 | -9.2 | 1.8 × 10-7 |
| L-10 | 324.11 | 312.15 | 346.291 | -1230.78 | -8.92 | -1.20 | 64.87 | 2.73 | -3.49 | -8.9 | 2.99 × 10-7 |
| L-11 | 362.95 | 355.03 | 358.390 | -978.70 | -8.75 | -0.54 | 68.24 | 5.78 | -1.73 | -9.3 | 1.52 × 10-7 |
| L-12 | 310.62 | 293.05 | 318.281 | -1097.51 | -9.10 | -0.63 | 63.15 | 6.08 | -2.90 | -9.7 | 7.75 × 10-8 |
| L-13 | 307.43 | 292.12 | 318.281 | -1085.40 | -8.70 | -0.81 | 63.21 | 3.28 | -4.21 | -9.7 | 7.75 × 10-8 |
| L-14 | 306.81 | 292.73 | 318.281 | -1039.91 | -8.76 | -0.95 | 63.28 | 2.26 | -3.78 | -9.0 | 2.53 × 10-7 |
| L-15 | 309.57 | 292.92 | 318.280 | -1037.21 | -8.92 | -0.85 | 63.23 | 6.99 | -4.21 | -9.4 | 1.29 × 10-7 |
| L-16 | 309.37 | 292.75 | 318.281 | -1074.80 | -9.11 | -0.53 | 63.10 | 5.77 | -4.21 | -9.2 | 1.8 × 10-7 |
| L-17 | 307.99 | 292.25 | 318.281 | -1096.81 | -9.27 | -0.68 | 63.05 | 6.35 | -3.36 | -9.4 | 1.29 × 10-7 |
| L-18 | 310.68 | 293.13 | 318.281 | -1075.81 | -9.46 | -0.70 | 63.09 | 4.36 | -4.21 | -9.8 | 6.54 × 10-8 |
| L-19 | 318.52 | 303.24 | 316.309 | -916.86 | -8.91 | -0.60 | 64.01 | 4.24 | -2.95 | -9.1 | 2.13 × 10-7 |
| L-20 | 339.22 | 321.24 | 342.303 | -797.93 | -9.28 | -0.78 | 65.43 | 5.07 | -3.78 | -9.2 | 1.8 × 10-7 |
| L-21 | 354.17 | 339.52 | 344.363 | -967.14 | -8.89 | -0.56 | 66.95 | 4.69 | -2.00 | -8.8 | 3.54 × 10-7 |
| L-22 | 342.47 | 322.54 | 330.336 | -945.44 | -8.88 | -0.60 | 65.58 | 5.27 | -2.30 | -9.3 | 1.52 × 10-7 |
| L-23 | 342.40 | 328.44 | 346.335 | -1126.65 | -9.10 | -0.63 | 66.02 | 6.00 | -3.69 | -8.8 | 3.54 × 10-7 |
| L-24 | 346.74 | 330.62 | 346.335 | -1074.31 | -8.86 | -0.42 | 66.20 | 6.87 | -3.65 | -8.6 | 4.96 × 10-7 |
| L-25 | 378.75 | 363.81 | 374.389 | -1205.32 | -8.87 | -0.45 | 68.90 | 4.70 | -3.19 | -9.5 | 1.09 × 10-7 |
| L-26 | 327.90 | 315.06 | 345.307 | -1075.35 | -9.30 | -0.98 | 64.97 | 8.29 | -4.53 | -9.2 | 1.8 × 10-7 |
| L-27 | 329.52 | 315.80 | 345.307 | -1063.75 | -8.79 | -0.93 | 65.14 | 2.03 | -4.14 | -9.1 | 2.13 × 10-7 |
| L-28 | 334.37 | 322.21 | 344.319 | -1105.44 | -9.23 | -0.78 | 65.52 | 8.44 | -3.37 | -9.1 | 2.13 × 10-7 |
| L-29 | 422.18 | 408.02 | 398.455 | -947.47 | -8.86 | -0.53 | 72.51 | 3.80 | -0.51 | -9.0 | 2.53 × 10-7 |
| L-30 | 321.70 | 305.60 | 347.279 | -926.60 | -9.24 | -1.23 | 64.29 | 9.24 | -5.56 | -9.3 | 1.52 × 10-7 |
| L-31 | 366.67 | 355.62 | 358.390 | -975.18 | -8.95 | -0.55 | 63.24 | 6.42 | -1.23 | -9.4 | 1.29 × 10-7 |
| L-32 | 379.22 | 368.78 | 378.380 | -786.89 | -8.95 | -0.65 | 69.33 | 4.70 | -2.56 | -10.3 | 2.81 × 10-8 |
| L-33 | 389.72 | 376.32 | 394.379 | -974.62 | -8.95 | -0.64 | 69.94 | 6.33 | -3.64 | -9.9 | 5.53 × 10-8 |
| L-34 | 401.87 | 384.52 | 410.378 | -1146.95 | -8.97 | -0.63 | 70.60 | 4.64 | -4.35 | -9.7 | 7.75 × 10-8 |
| L-35 | 305.22 | 288.71 | 316.265 | -922.52 | -8.92 | -1.35 | 63.01 | 4.40 | -2.95 | -9.1 | 2.13 × 10-7 |
| L-36 | 341.92 | 328.44 | 346.335 | -1125.55 | -9.46 | -0.75 | 65.97 | 3.16 | -3.73 | -9.2 | 1.8 × 10-7 |
| L-37 | 351.81 | 330.21 | 346.335 | -1104.84 | -8.85 | -0.57 | 66.21 | 6.61 | -3.36 | -9.6 | 9.17 × 10-8 |
| L-38 | 336.55 | 343.60 | 360.362 | -772.15 | -8.4 | -1.35 | 67.58 | 4.24 | -3.32 | -8.7 | 4.19 × 10-7 |
| L-39 | 355.00 | 344.88 | 360.362 | -1158.51 | -8.92 | -0.39 | 67.34 | 5.60 | -3.47 | -9.4 | 1.29 × 10-7 |
| L-40 | 384.66 | 370.00 | 378.380 | -781.27 | -8.96 | -0.63 | 69.42 | 4.90 | -2.19 | -9.4 | 1.29 × 10-7 |
| L-41 | 379.14 | 368.78 | 378.380 | -786.74 | -8.95 | -0.64 | 69.33 | 7.16 | -2.56 | -9.5 | 1.09 × 10-7 |
| L-42 | 376.42 | 368.490 | 378.380 | -788.10 | -8.92 | -0.56 | 69.29 | 4.42 | -2.56 | -10.1 | 3.94 × 10-8 |
| L-43 | 378.23 | 368.76 | 378.380 | -792.57 | -9.26 | -0.73 | 69.28 | 4.32 | -2.56 | -10.3 | 2.81 × 10-8 |
| L-44 | 378.02 | 368.77 | 378.380 | -795.72 | -8.88 | -0.53 | 69.32 | 5.27 | -2.56 | -10.0 | 4.67 × 10-8 |
| L-45 | 376.69 | 368.68 | 378.380 | -766.29 | -8.87 | -0.61 | 69.33 | 5.08 | -1.37 | -9.1 | 2.13 × 10-7 |
| L-46 | 317.80 | 303.40 | 316.309 | -910.75 | -8.91 | -0.60 | 64.03 | 4.44 | -2.56 | -8.7 | 4.19 × 10-7 |
| L-47 | 321.47 | 304.01 | 316.309 | -884.14 | -8.61 | -0.43 | 64.10 | 2.90 | -2.95 | -9.0 | 2.53 × 10-7 |
| L-48 | 322.50 | 304.19 | 316.309 | -924.29 | -8.88 | -0.60 | 64.10 | 5.14 | -2.78 | -9.4 | 1.29 × 10-7 |
| L-49 | 317.83 | 303.07 | 316.309 | -928.79 | -8.86 | -0.60 | 64.01 | 4.15 | -2.95 | -9.1 | 2.13 × 10-7 |
| L-50 | 314.49 | 296.42 | 317.297 | -886.04 | -9.02 | -0.60 | 63.43 | 6.15 | -3.74 | -9.3 | 1.52 × 10-7 |
| L-51 | 314.68 | 296.22 | 317.297 | -886.91 | -8.95 | -0.66 | 63.45 | 4.18 | -4.84 | -9.4 | 1.29 × 10-7 |
| L-52 | 314.94 | 296.33 | 317.297 | -886.91 | -9.07 | -0.66 | 63.43 | 7.70 | -4.84 | -9.6 | 9.17 × 10-8 |
| L-53 | 312.51 | 295.83 | 317.297 | -886.25 | -8.73 | -0.56 | 63.44 | 4.53 | -4.84 | -9.0 | 2.53 × 10-7 |
| L-54 | 310.73 | 295.35 | 317.297 | -901.51 | -8.37 | -0.61 | 63.50 | 5.91 | -4.84 | -9.3 | 1.52 × 10-7 |
| L-55 | 314.22 | 296.60 | 317.297 | -857.94 | -8.71 | -0.79 | 63.56 | 3.14 | -4.16 | -9.2 | 1.8 × 10-7 |
| L-56 | 334.65 | 323.91 | 344.319 | -1016.87 | -8.86 | -0.67 | 65.72 | 4.48 | -3.81 | -8.9 | 2.99 × 10-7 |
| L-57 | 327.44 | 310.83 | 332.308 | -1061.47 | -8.70 | -0.85 | 64.73 | 5.08 | -4.03 | -8.9 | 2.99 × 10-7 |
| L-58 | 319.90 | 300.69 | 334.28 | -1274.99 | -9.53 | -0.87 | 63.72 | 5.50 | -3.98 | -9.6 | 9.17 × 10-8 |
| L-59 | 314.13 | 262.22 | 332.264 | -1103.87 | -9.42 | -1.43 | 63.52 | 2.42 | -4.04 | -9.3 | 1.52 × 10-7 |
| L-60 | 328.59 | 312.74 | 362.294 | -1314.32 | -7.26 | 1.70 | 64.63 | 2.91 | -4.60 | -9.7 | 7.75 × 10-8 |
| L-61 | 382.20 | 371.96 | 392.363 | -820.40 | -9.21 | -1.40 | 69.70 | 3.03 | -2.39 | -9.9 | 5.53 × 10-8 |
| L-62 | 397.56 | 386.77 | 392.407 | -807.24 | -8.93 | -0.64 | 70.79 | 4.96 | -2.38 | -9.4 | 1.29 × 10-7 |
| L-63 | 412.39 | 400.20 | 421.405 | -950.65 | -8.78 | -0.92 | 71.99 | 2.43 | -3.20 | -9.9 | 5.53 × 10-8 |
| L-64 | 409.23 | 398.84 | 421.405 | -965.63 | -9.22 | -0.72 | 71.72 | 5.75 | -3.96 | -9.0 | 2.53 × 10-7 |
| L-65 | 382.23 | 373.66 | 403.387 | -1381.02 | -9.02 | -0.54 | 69.69 | 7.88 | -4.88 | -10.0 | 4.67 × 10-8 |
| L-66 | 419.99 | 405.97 | 437.404 | -1190.20 | -8.93 | -0.73 | 72.37 | 7.57 | -4.68 | -11.2 | 6.16 × 10-9 |
| L-67 | 429.09 | 420.51 | 455.487 | -1080.32 | -8.60 | -0.81 | 73.65 | 3.20 | -5.30 | -9.8 | 6.54 × 10-8 |
| L-68 | 445.01 | 429.70 | 454.499 | -1017.46 | -8.84 | -0.61 | 74.29 | 4.70 | -3.19 | -10.0 | 4.67 × 10-8 |
| L-69 | 314.13 | 296.22 | 332.264 | -1103.87 | -9.42 | -1.43 | 63.52 | 2.42 | -4.04 | -9.2 | 1.8 × 10-7 |
| L-70 | 408.37 | 389.88 | 429.375 | -1161.50 | -9.24 | -0.91 | 71.04 | 7.34 | -2.79 | -10.1 | 3.94 × 10-8 |
| L-71 | 467.25 | 457.54 | 513.333 | -692.48 | -9.26 | -0.92 | 76.52 | 4.06 | -0.41 | -9.9 | 5.53 × 10-8 |
| L-72 | 359.61 | 349.35 | 382.328 | -739.91 | -9.28 | -1.53 | 67.88 | 8.13 | -3.81 | -9.6 | 9.17 × 10-8 |
| L-73 | 420.87 | 406.27 | 462.285 | -865.88 | -9.03 | -0.82 | 72.40 | 5.41 | -2.36 | -10.4 | 2.38 × 10-8 |
| L-74 | 401.00 | 388.51 | 429.375 | -1181.82 | -8.85 | -0.86 | 71.00 | 2.95 | -3.16 | -10.4 | 2.38 × 10-8 |
| L-75 | 359.65 | 343.68 | 391.307 | -1248.52 | -9.15 | -1.42 | 67.43 | 7.18 | -3.27 | -9.8 | 6.54 × 10-8 |
| L-76 | 400.15 | 391.35 | 444.416 | -1196.64 | -9.08 | -1.50 | 71.33 | 8.89 | -3.87 | -8.4 | 6.95 × 10-7 |
| L-77 | 374.41 | 367.41 | 387.388 | -994.18 | -8.78 | -1.34 | 69.42 | 2.69 | -3.28 | -9.5 | 1.09 × 10-7 |
| L-78 | 393.53 | 386.81 | 424.429 | -811.23 | -9.26 | -2.16 | 71.08 | 4.28 | -1.28 | -8.3 | 8.23 × 10-7 |
| L-79 | 397.02 | 384.84 | 430.359 | -1390.07 | -9.27 | -0.80 | 70.59 | 4.24 | -1.68 | -9.3 | 1.52 × 10-7 |
| L-80 | 384.17 | 373.79 | 389.404 | -1178.49 | -9.46 | -0.61 | 69.68 | 3.45 | -3.56 | -8.1 | 1.15 × 10-6 |
| L-81 | 377.49 | 356.44 | 390.344 | -1433.12 | -9.12 | -0.63 | 68.29 | 7.53 | -3.01 | -9.9 | 5.53 × 10-8 |
| L-82 | 371.36 | 362.32 | 405.359 | -1405.69 | -9.23 | -0.77 | 68.77 | 7.35 | -5.08 | -9.9 | 5.53 × 10-8 |
| L-83 | 365.25 | 377.58 | 417.370 | -1446.00 | -9.22 | -0.83 | 70.02 | 4.08 | -4.83 | -10.3 | 2.81 × 10-8 |
| L-84 | 416.05 | 398.50 | 422.389 | -1020.86 | -8.84 | -1.29 | 71.92 | 4.15 | -1.43 | -10.2 | 3.33 × 10-8 |
| L-85 | 362.57 | 343.35 | 380.780 | -1169.65 | -9.22 | -0.71 | 67.22 | 6.09 | -2.01 | -10.0 | 4.67 × 10-8 |
| L-86 | 351.05 | 334.85 | 381.768 | -1143.72 | -9.24 | -0.86 | 66.56 | 7.06 | -3.18 | -9.0 | 2.53 × 10-7 |
| L-87 | 356.75 | 341.14 | 375.333 | -1273.51 | -8.86 | -0.87 | 67.16 | 3.20 | -3.73 | -10.1 | 3.94 × 10-8 |
| L-88 | 441.77 | 432.80 | 506.294 | -1222.23 | -8.89 | -0.98 | 74.62 | 2.60 | -1.43 | -9.1 | 2.13 × 10-7 |
| L-89 | 414.10 | 395.43 | 431.397 | -1480.28 | -8.91 | -0.97 | 71.58 | 1.71 | -4.24 | -9.9 | 5.53 × 10-8 |
| L-90 | 374.56 | 354.44 | 429.303 | -1980.61 | -9.61 | -1.07 | 68.11 | 5.16 | -3.32 | -11.0 | 8.63 × 10-9 |
| L-91 | 366.82 | 353.18 | 429.303 | -1883.43 | -8.97 | -1.34 | 68.22 | 2.20 | -4.61 | -10.6 | 1.7 × 10-8 |
| L-92 | 383.87 | 377.83 | 478.668 | -1304.22 | -8.93 | -1.29 | 70.22 | 2.39 | -3.86 | -10.6 | 1.7 × 10-8 |
| L-93 | 346.88 | 331.86 | 376.321 | -1278.09 | -8.27 | -0.95 | 66.56 | 9.80 | -6.46 | -9.8 | 6.54 × 10-8 |
| L-94 | 355.27 | 336.19 | 362.334 | -1345.06 | -9.05 | -0.62 | 66.66 | 5.98 | -3.44 | -9.6 | 9.17 × 10-8 |
| L-95 | 348.99 | 336.74 | 395.751 | -1257.37 | -9.55 | -1.10 | 66.70 | 3.19 | -4.66 | -10.8 | 1.21 × 10-8 |
| L-96 | 348.97 | 331.79 | 382.752 | -1347.49 | -9.61 | -0.99 | 66.26 | 3.61 | -3.26 | -10.2 | 3.33 × 10-8 |
| L-97 | 463.02 | 462.15 | 554.766 | -1285.57 | -8.97 | -1.01 | 76.98 | 2.34 | -3.32 | -9.5 | 1.09 × 10-7 |
| L-98 | 354.89 | 338.44 | 393.372 | -1316.26 | -8.41 | -0.90 | 67.05 | 5.09 | -4.81 | -10.1 | 3.94 × 10-8 |
| L-99 | 367.46 | 348.00 | 391.332 | -1431.91 | -8.83 | -1.08 | 67.77 | 2.56 | -6.13 | -10.5 | 2.01 × 10-8 |
| L-100 | 448.28 | 440.64 | 465.458 | -1246.87 | -9.16 | -0.74 | 75.13 | 5.57 | -4.45 | -9.7 | 7.75 × 10-8 |
In the study, the docking results of Hesperetin and its derivatives were found in the range of -8.7 to -11.0 kcal.mol-1, while the binding affinity of the drug Fluconazole was found to be -8.6 kcal.mol-1. This change in binding energy means that the affinity of the molecule under study is increased approximately 100 times. In particular, it was observed that the -CONH2 substituent attached to the R1 and R2 positions of the substituted groups contributed positively to the affinity. It was also observed that the increase in the area and volume values of the molecules was in parallel with the increase in the binding energies of the molecules. In order to make more detailed analysis with the obtained interaction map, the active site amino acids interacting with the ligands are organized in Table 5 by indicating the distances of the interaction types.
Table 5. Ligand-amino acid interactions.
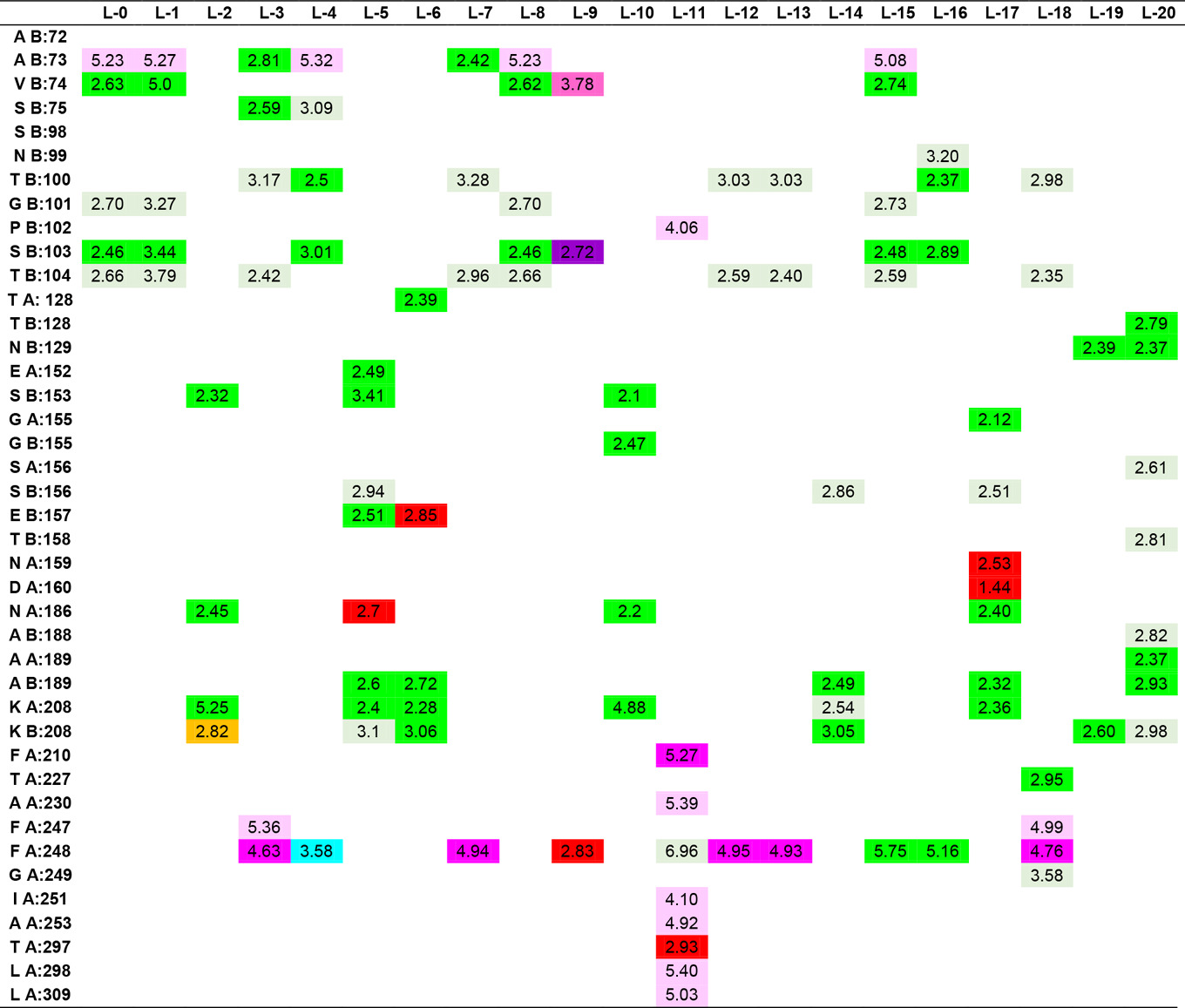  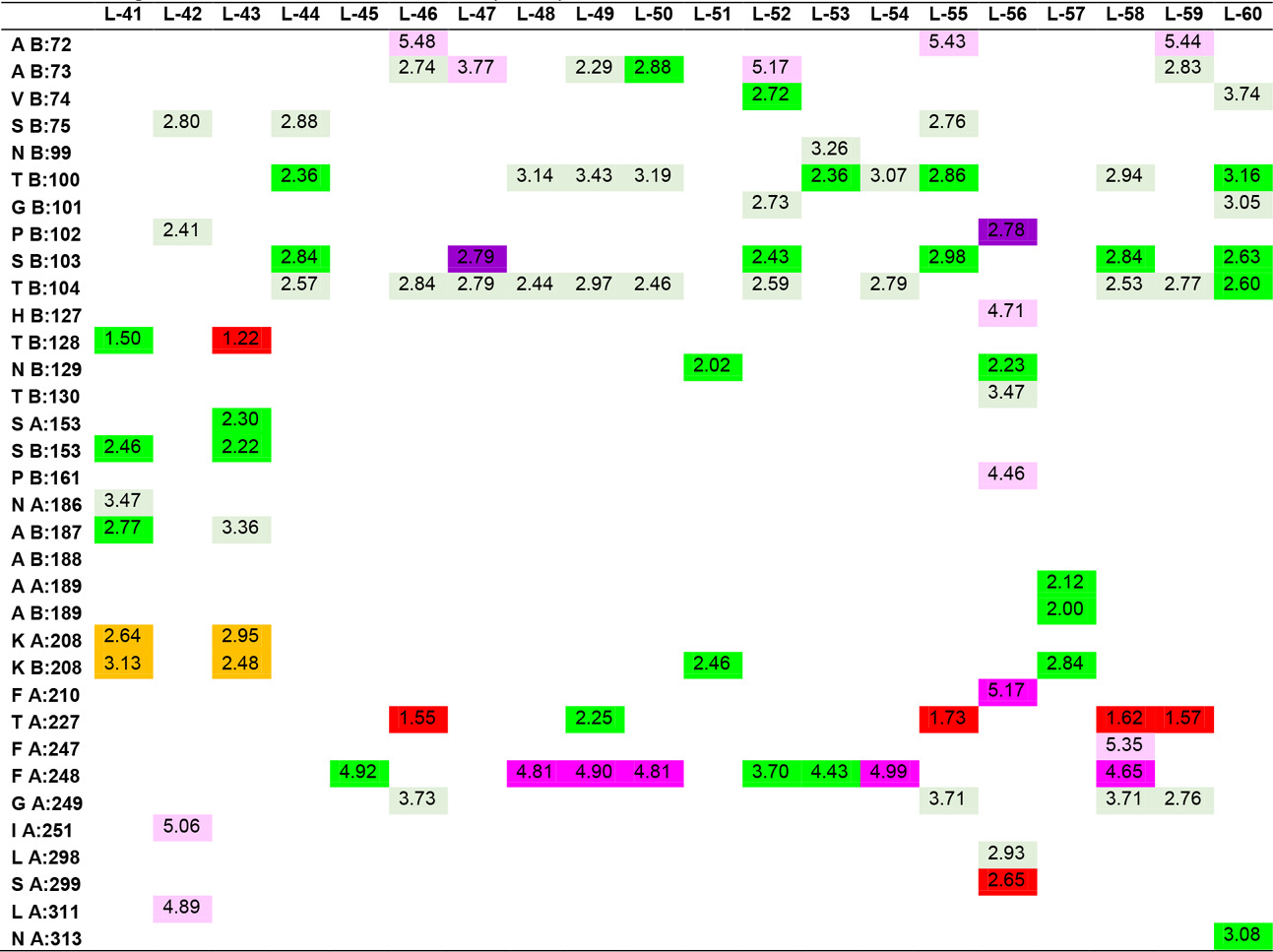 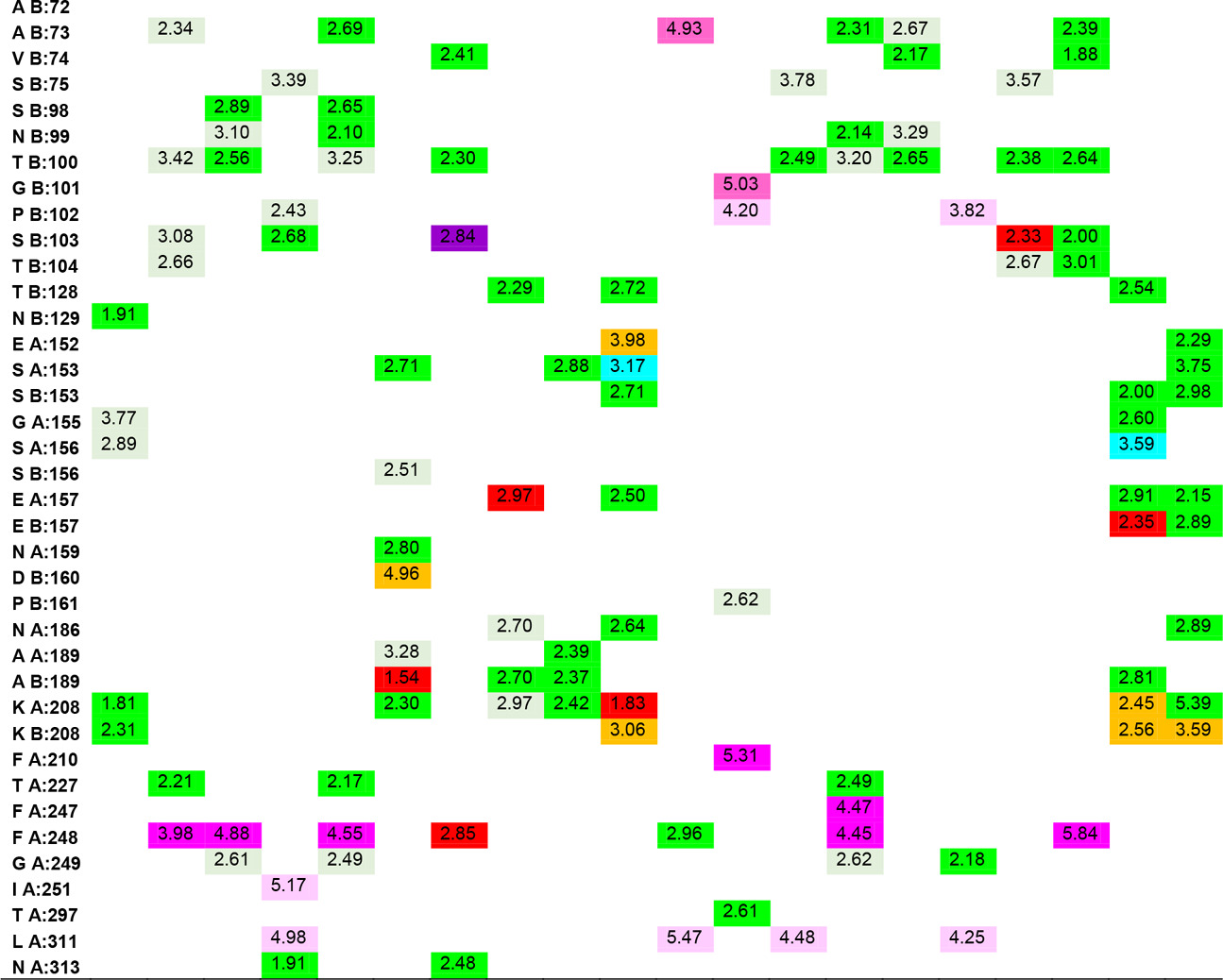 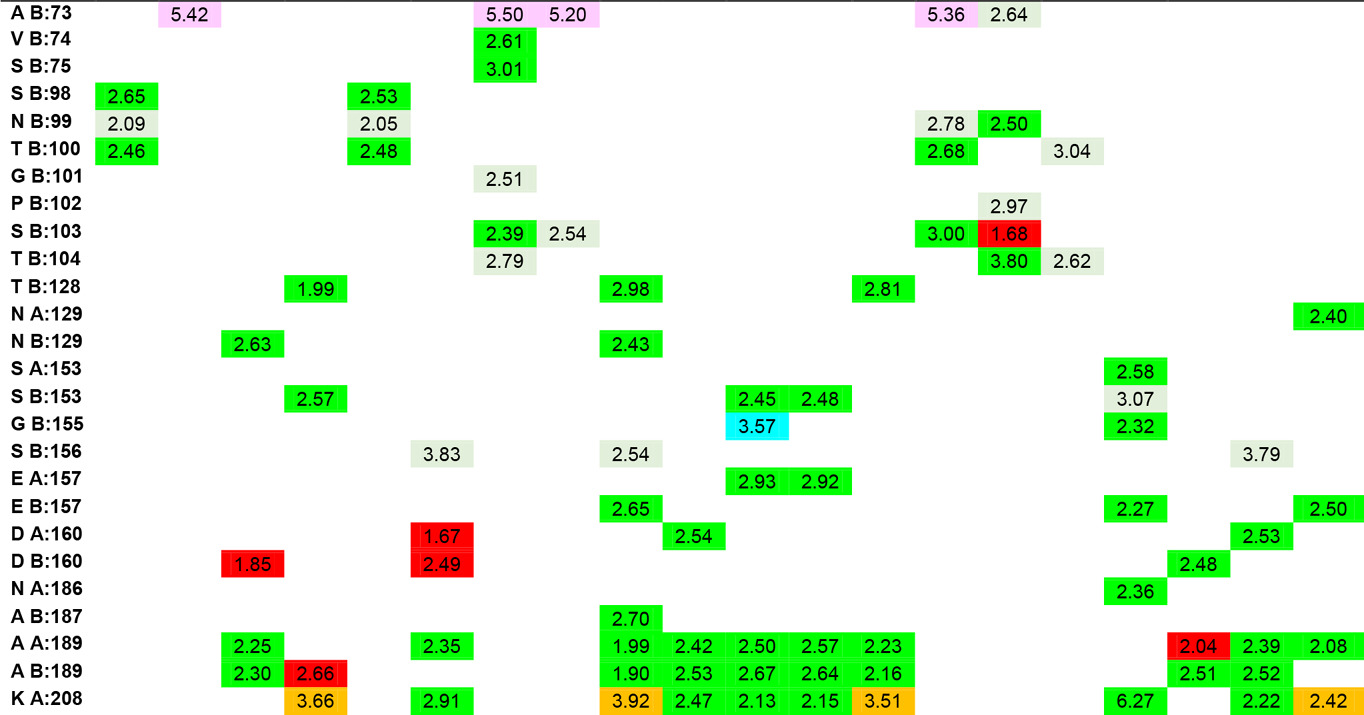  |
In addition to the results, it can be said that the derivatizations made at the R4, R7 and R8 positions contribute less to the affinity than those made at the R1 and R5 ends, even if there is an increase in the binding energy, they support unfavorable donor-donor interactions. The R6 position had no positive or negative effect on affinity. Especially -NH2, -OH and -CH3 groups can be said to increase protein-ligand interactions by increasing the number of conventional hydrogen bonds which are very important in intermolecular interactions. When the interactions of Hesperetin molecule with the target macromolecule structure are considered, it is seen that it interacts with SeR103 and Val74 amino acids with conventional hydrogen bonds of 2.46 and 2.63 Å, respectively, and with Gly101 and ThR104 amino acids with carbon-hydrogen bonds of 2.70 and 2.66 Å, respectively also. A remarkable point in the interaction map is that the interacting amino acids belong to the B lobe. As a result of the substituted groups attached to the molecule in the derivatization study, interactions with amino acids in the A lobe could also be observed. Therefore, this allows us to interpretation of both the effector molecules designed and the active site characterization of the protein structure. The critical amino acids in the active site of the protein were determined by analyzing the data in Table 5. As a result of these examinations, the amino acids that interacted with the closest conventional hydrogen bond were determined as Ala B: 73, Thr B: 100, Ser B: 103, Thr B: 128, Asn B: 129, Ser B: 150, Ala A: 189, Ala B: 189, Lys A: 208. Therefore, it can be said that a possible mutation in these amino acids is critical in protein structure and function. In the analyzed results, the proposed molecules are ligands L-66, L-92, L-95, L-90 and L-99.
Conclusions
The aim of this study was to design new drug molecules that can show antifungal effect against C. glabrata. Out of 100 designed molecules, 5 candidate active molecules were selected. This is because there is more than one eliminating factor. While candidate molecules are expected to have high binding energies, they are also expected to interact in close proximity with the critical active site amino acids in the targeted protein structure. Another criterion evaluated is the type and characteristics of the observed interactions. At the same time, in the results obtained, the amino acids in the active site of the protein and determining the interactions with the molecules were generally polar and uncharged amino acids. Exceptionally, the apolar and aliphatic amino acid alanine and the positively charged amino acid lysine were also significant. Considering that C. glabrata develops a mechanism of resistance to drugs by mutating in a rapid period of time, it is necessary to design new candidate molecules that are antifungal effective, resistant to mutations and highly selective. It is recommended to further improve the information obtained on the inhibition potential of candidate molecules by targeting different protein structures of the organism in future studies.